Figure 3.
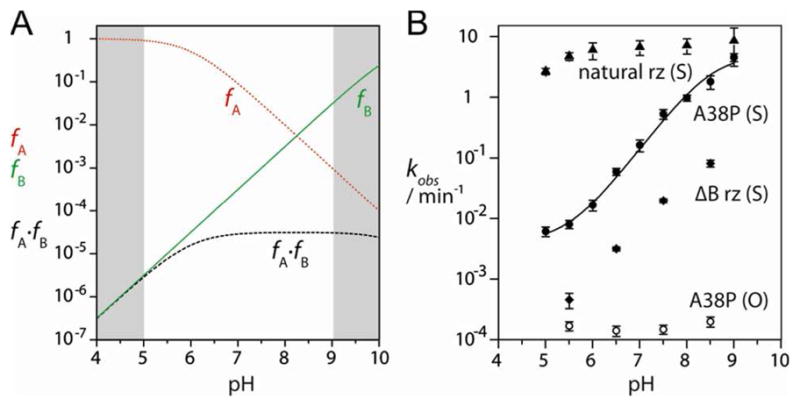
The pH dependence of cleavage rates for the 5′-PO and 5′-PS substrate with various ribozyme constructs.
A. Simulation of the reaction rate as a function of pH for a general acid of pKa = 6 and a general base of pKa = 10.5. The fractions of the protonated acid (fA – red dotted line), unprotonated base (fB- green solid line) and fA × fB (dashed line) are plotted as a function of pH. In all simulations, the shaded regions lie outside the experimentally accessible range of pH; note that the reduction in fA × fB at high pH falls outside the observable region. If general acid catalysis is not required, the pH dependence should simply follow fB.
B. Experimental cleavage rates for the 5′-PO and 5′-PS substrate with various ribozyme constructs are plotted on a logarithmic scale as a function of pH. Filled triangles, 5′-PS substrate + natural-sequence ribozyme; filled circles, 5′-PS substrate + A38P ribozyme; filled diamonds, 5′-PS substrate + ΔB ribozyme (lacking loop B); open circles, 5′-PO substrate + A38P ribozyme. The data for the 5′-PS substrate + A38P ribozyme have been fitted to a single ionization model with additional pH-independent component as discussed in the text, yielding an apparent pKa of 8.5± 0.2.
