Figure 1. The PBcR single-molecule read correction and assembly pipeline.
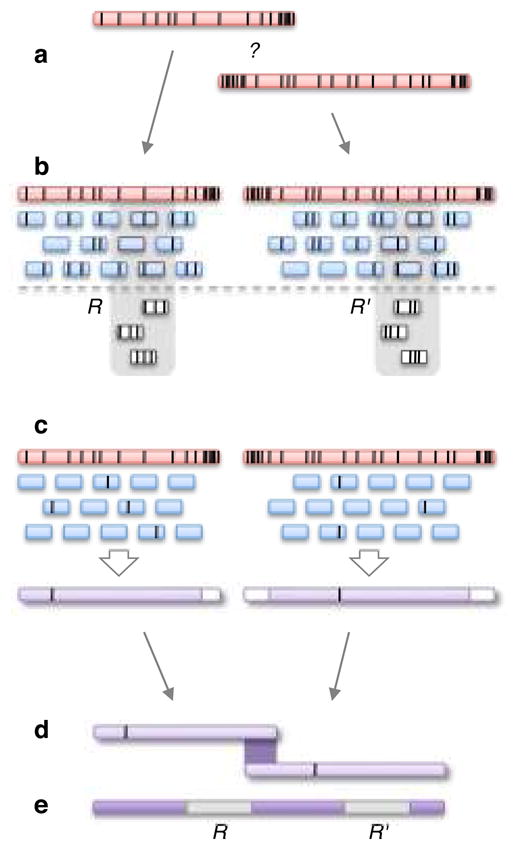
a) The high-error, indicated by black vertical bars, in single-pass PacBio RS sequences obscures overlaps. b) Given a high-accuracy sequence (~99% identical to the truth), the error between it and a PacBio RS sequence is half the error between two PacBio RS sequences. Therefore, accurate alignments can be computed. In this example, black bars in the short-reads indicate “mapping errors” that are a combination of the sequencing error in both the long and short reads. In addition, a two-copy inexact repeat is present (outlined in gray) leading to “pileups” of reads at each copy. To avoid mapping reads to the wrong repeat copy, the pipeline selects a cutoff, C, and only the top C hits for each short read are used. The spurious mappings (in white) are discarded. c) The remaining alignments are used to generate a new consensus sequence, trimming and splitting long reads whenever there is a gap in the short-read tiling. Sequencing errors, indicated in black, may propagate to the PBcR read in rare cases where sequencing error co-occurs. d) After correction, overlaps between long PBcR sequences can be easily detected. e) The resulting assembly is able to span repeats that are unresolvable using only the short reads.
