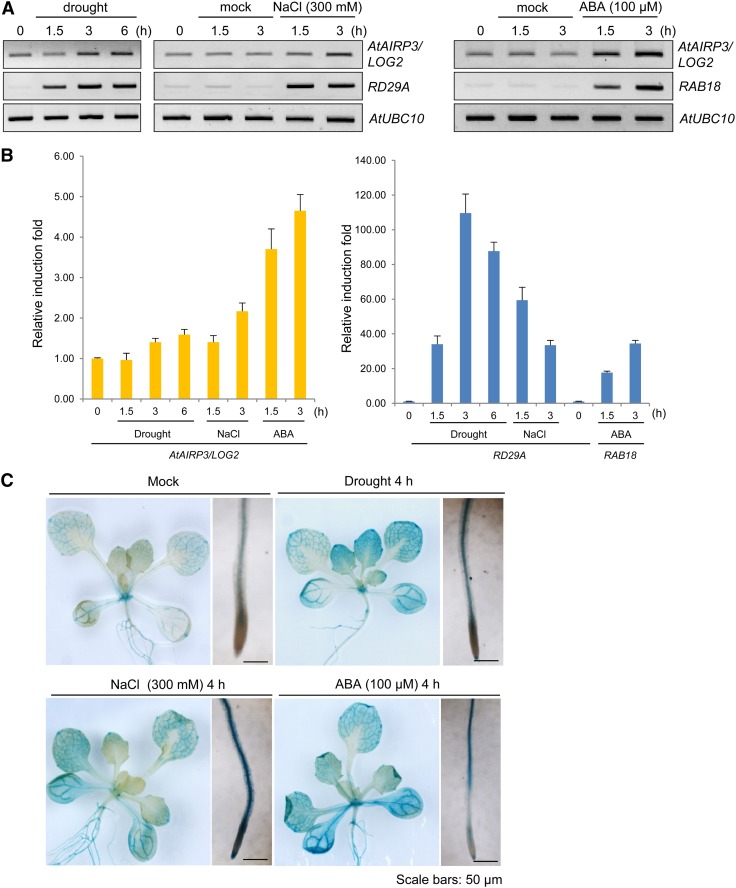
Figure 2.
Induction of AtAIRP3/LOG2 in response to drought, high salinity, and ABA. A, RT-PCR analysis of AtAIRP3/LOG2. Light-grown, 10-d-old Arabidopsis seedlings were subjected to drought (0, 1.5, 3, and 6 h), 300 mm NaCl (0, 1.5, and 3 h), or 100 μm ABA (1.5 and 3 h). Total RNA prepared from the treated tissues was analyzed by RT-PCR using gene-specific primer sets. RD29A served as a positive control for drought and salt treatments, whereas RAB18 served as a positive control for ABA induction. AtUBC10 was used as a loading control. B, Real-time qRT-PCR analysis of AtAIRP3/LOG2. Total RNA was isolated from the treated tissues and used for real-time qRT-PCR. Fold induction of AtAIRP3/LOG2 was normalized to levels of glyceraldehyde-3-phosphate dehydrogenase C subunit mRNA, which served as an internal control. Bars indicate means ± sd from three independent experiments. C, AtAIRP3/LOG2 promoter-GUS assay. Light-grown, 10-d-old AtAIRP3-promoter:GUS transgenic T3 plants were grown under drought conditions (4 h) with 300 mm NaCl (4 h) or with 100 μm ABA (4 h). Histochemical localization of GUS activities in leaf and root tissues was visualized by 5-bromo-4-chloro-3-indolyl β-D-glucoside staining for 1 h. Bars = 50 μm. [See online article for color version of this figure.]