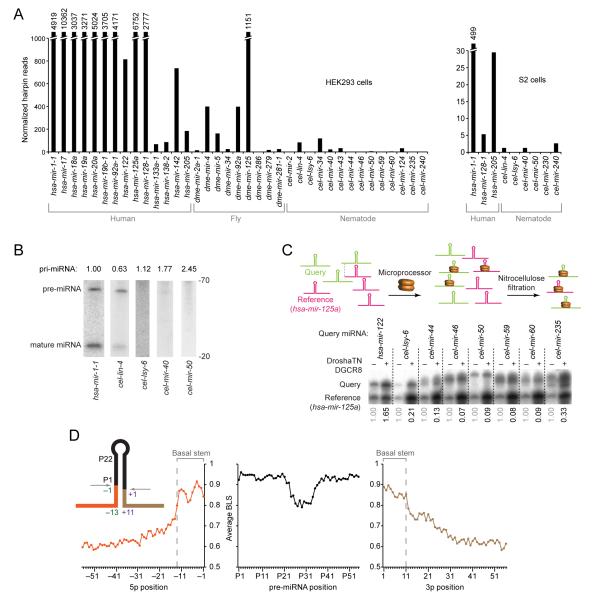
Figure 1. Existence of Unknown Features Specifying Human Pri-miRNAs.
(A) Processing of human, fly, and nematode pri-miRNAs in human cells and Drosophila cells. Cells were transfected with plasmids expressing the indicated pri-miRNA hairpins with ~100 flanking genomic nucleotides on each side of each hairpin (Figure S1A), and total RNA was pooled for small-RNA sequencing. Plotted are small-RNA reads derived from the indicated pri-miRNAs.
(B) Accumulation of pri-miRNA, pre-miRNA and miRNA after expressing the indicated pri-miRNAs in HEK293T cells. Pre-miRNA and mature species were measured by RNA blot of total RNA from cells transfected with plasmids expressing the indicated pri-miRNA (full gel images, including in vitro transcribed cognate positive controls, in Figure S1B). Relative pri-miRNA levels (indicated above the lanes) are from ribonuclease protection assays, normalized to the signals for neomycin phosphotransferase mRNA also expressed from each expression plasmid.
(C) Relative binding of C. elegans and human pri-miRNAs to the Microprocessor. In the competitive binding assay (top, schematic), radiolabeled query pri-miRNA was mixed with the radiolabeled shorter reference pri-miRNA (human mir-125a) and incubated in excess over catalytically impaired Drosha (Drosha-TN) and DGCR8. Bound RNA was filtered on nitrocellulose and eluted for analysis on a denaturing gel. Phosphorimaging (bottom) indicated the relative amounts of input (−) and bound (+) RNAs. Numbers below each lane indicate the ratio of bound query to bound reference pri-miRNAs, normalized to their input ratio.
(D) Nucleotide conservation of human pri-miRNAs conserved to mouse, reported as the average branch-length score (BLS) at each position. Positions are numbered based on the inferred Drosha cleavage site (inset); negative indices are upstream of the 5p Drosha cleavage site, indices with “P” count from the 5′ end of the pre-miRNA, and positive indices are downstream of the 3p Drosha cleavage site.