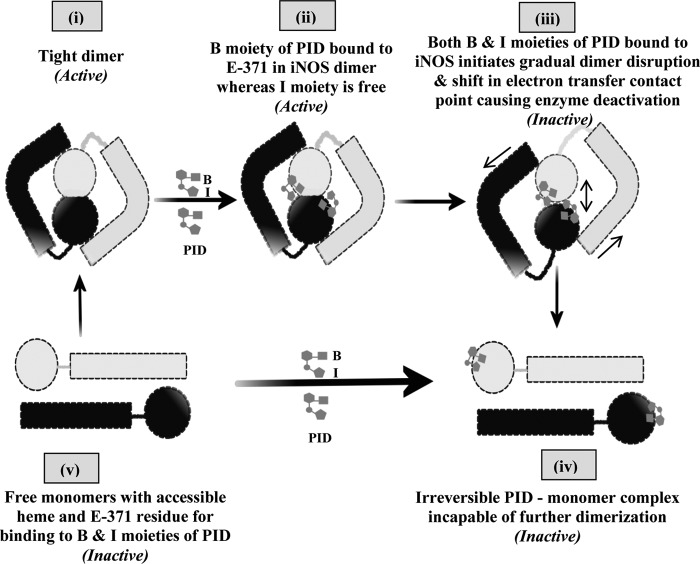
FIGURE 8.
Scheme for PID-induced inhibition of iNOS. The scheme pictorially describes the mode of inhibition of iNOS by PID as well as the fate of the iNOS monomer and dimer after PID binding. In the proposed model, PID binds to the Glu-371 (E-371) residue of the heme containing iNOS monomer (v) through its benzodioxole moiety (B) as well as the free sixth axial position of the heme iron through its imidazole moiety (I) almost simultaneously and forms an irreversible iNOS-monomer-PID complex (iv). However, in the iNOS dimer (i), the benzodioxole moiety of PID initiates binding to the open Glu-371 residue and consequently disturbs helix α7a and thereafter helix 8 that form the critical part of the dimer interface (ii). This apparently helps to slowly open up the heme distal pocket to provide direct access for the unbound imidazole moiety of the already iNOS-Glu-371-bound PID to the iNOS heme (that was previously sequestered), which on coordinating with the sixth axial position of the heme iron initiates dimer separation (iii) and apparently causes loss of effective contact between the reductase and oxygenase domains and abrogation of the functional inter-domain electron transfer and deactivation of the enzyme, even when the dimers are not fully parted. This subsequently leads to the irreversible monomerization of the enzyme through complete steric separation of the two monomeric subunits constituting the iNOS dimer and formation of an irreversible iNOS-monomer-PID complex (iv) that is not recoverable to the active dimer even with high concentrations of H4B and Arg.