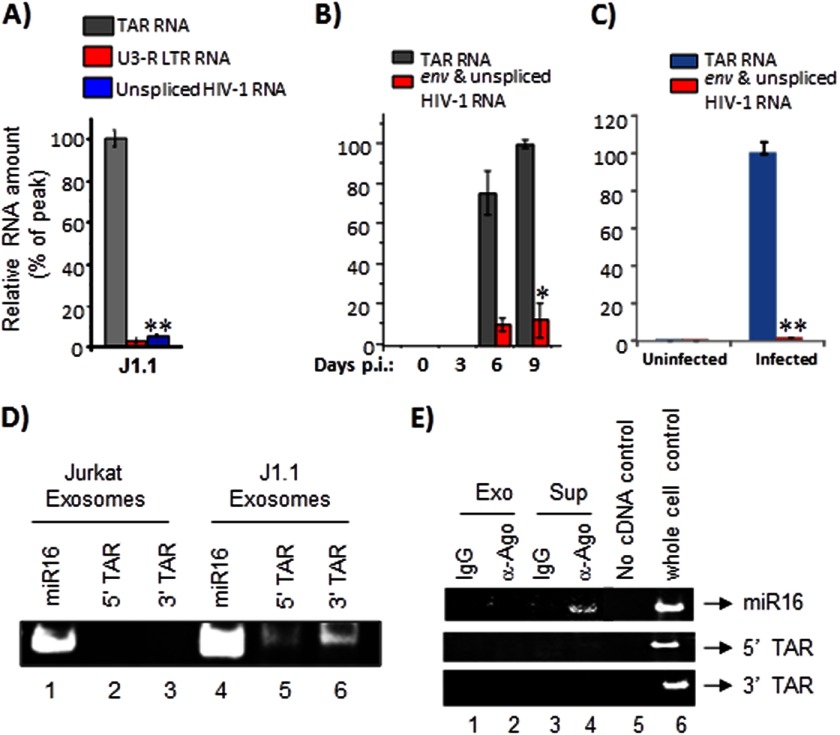
FIGURE 3.
Exosomes derived from HIV-1-infected cells contain TAR RNA. A, total RNA was isolated from J1.1-derived exosomes and analyzed by qRT-PCR with primers specific for TAR RNA, U3-R LTR sequence, and unspliced HIV-1 RNA as described in A. Error bars show the standard deviation from three independent RNA preparations; double asterisk indicates p ≤ 0.01. B, primary cells from a healthy donor (PBMCs) were activated with anti-CD3/CD28 and then infected with HIV-1IIIB (ABI). Culture supernatants were harvested in 3, 6, and 9 days after inoculation; exosomal fraction was separated, and then the total RNA was purified and analyzed by quantitative RT-PCR with primers specific for TAR and unspliced HIV-1 RNA. Results are presented as a mean of three independent measurements ±S.D.; the asterisk indicates p ≤ 0.05. C, primary cells were activated as described in B. When 10–15% of the cells were infected, the culture was harvested, washed, and incubated for 7 days in complete medium (exosome-free) supplemented with 1 ng/ml IL-7 (R&D Systems) to allow the cells to revert to quiescence. Uninfected cells were maintained alongside as controls. Total RNA was obtained from exosomes enriched from culture supernatants of uninfected and infected cells and analyzed by qRT-PCR with primers for TAR and env RNA. Error bars show the standard deviation from three independent preparations. Double asterisk indicates p ≤ 0.01. D, total RNA was isolated from Jurkat and J1.1-derived exosomes and analyzed by RT-PCR (QuantiMiR small RNA analysis method) using primers specific to miR16, 5′, and 3′ TAR miRNA. After the RT-PCR, the amplified products were resolved on a 4–20% Tris-glycine polyacrylamide gel and visualized after staining with ethidium bromide. E, Jurkat and J1.1-derived exosomes were immunoprecipitated with anti-Ago2 antibodies. Immunoprecipitations performed with IgG antibodies were used as controls. All RNA associated with the antibodies was extracted and utilized for cDNA synthesis using the QuantiMiR small RNA isolation and analysis protocol. Half of the isolated RNA was utilized in the cDNA amplification procedure, and 30% of the amplified cDNA was used in the RT-PCR with 5′ primers against miR16, 5′ TAR miRNA, and 3′ TAR miRNA. The amplified DNA products were resolved in a polyacrylamide gel, stained with ethidium bromide, and visualized using a Molecular Imager ChemiDoc XRS system (Bio-Rad). Exo, exosomes; Sup, culture supernatant.