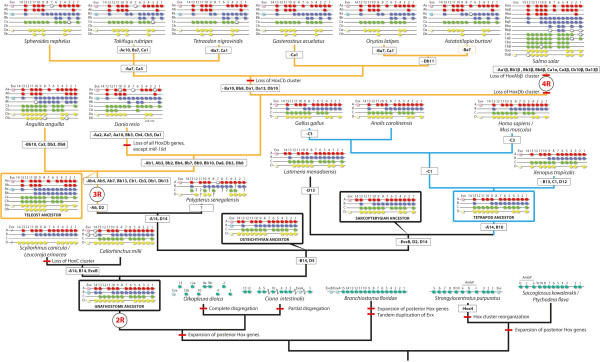
Figure 2.
Reconstructed evolution of Hox gene families within deuterostomes. The Hox genes and clusters of those representative species with complete or almost complete Hox cluster sequences are shown, and gene losses (thin black squares) or other events (crossed red lines), such as Hox cluster duplication or loss are inferred. The ancestral conditions are reconstructed taking into account the information of species with non-complete Hox cluster sequences (see the main text). Pre-duplicative clusters are shown in turquoise; vertebrate Hox clusters are type-coloured: red, HoxA; blue, HoxB; green, HoxC; yellow, HoxD. For the sake of clarity, the phylogenetic relationships of tetrapods are shown in light blue and those of teleosts in orange. White squares indicate pseudogenes. Evx genes are shown when possible, with lighter colours. 2R, 3R and 4R indicate two, three teleost-specific and four salmonid-specific rounds of whole genome duplication, respectively. The phylogenetic relationships of teleosts here are based on [98].