Figure 4.
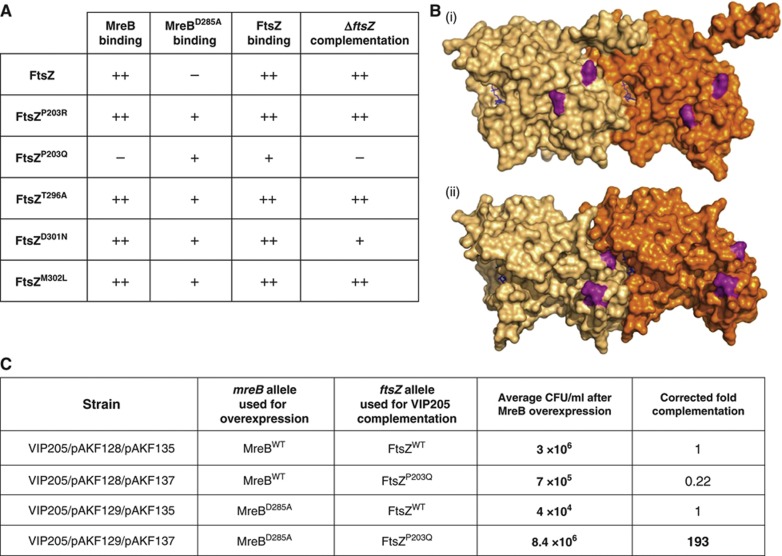
Single aa changes in FtsZ restore functional interaction with MreBD285A. (A) Table summarising the phenotypes of ftsZ point mutants identified in the ftsZ–mreBD285A PCR mutagenesis screen. Bacterial two-hybrid (BTH) scores represent signal strength from BTH101 double transformations, plated on NA plates with selective antibiotics and X-gal (see Supplementary Figure S5A for images of plates); ++=strong binding signal,+=weaker binding signal, −=no detectable signal. Details of plasmids used in these assays can be found in Supplementary Table SI. ‘ΔftsZ complementation scores’ are representative of CFU measurements shown in Supplementary Figure S5B; ++=full (comparable to ftsZwt complementation control),+=partial (100 drop in CFU compared to ftsZwt control) and −=no complementation (same CFU as −ve empty vector contol). Complementation was assayed using the VIP205 (MC1061: Ptac::ftsZ) strain complementing with Ptet::ftsZ (pAKF135) constructs containing the identified point mutations (pAKF136-140), the full list of plasmids can be found in Supplementary Table SI. Note that only the FtsZP203R variant has no detectable MreB binding and cannot complement the VIP205 ftsZ depletion strain. (B) Structural model of an FtsZ dimer of M. jannaschii (i) and S. aureus (ii) (Oliva et al, 2004; Matsui et al, 2012) showing the positions of the aas in FtsZ (purple), changes to which can restore the interaction with MreBD285A. The two monomers of the FtsZ dimer are shown in different shades of orange. GTP molecules are shown in blue. Identification of the equivalent aa positions in E. coli FtsZ were based on a sequence alignment of FtsZ (not shown). (C) FtsZP203Q can rescue MreBD285A expression. E. coli VIP205 (Ptac::ftsZ) cells were grown without IPTG inducer giving FtsZ depletion. This was complemented by expression of FtsZ from pAKF135 (Ptet::ftsZ) or pAKF137 (Ptet::ftsZP203Q) induced using 20 ng/ml chlorotetracycline. A FtsZ depletion system was used in this assay as overexpression of ftsZ can lead to cell elongation and thus CFU reduction. Strains contained either pAKF128 (PBAD::mreBCD) or pAKF129 (PBAD::mreBD285ACD) induced with 0.005% arabinose for controlled MreB overexpression. Cells were grown in M9 media for 8 h, serially diluted 1/10 and used for enumeration on NA plates with antibiotics and 30 μM IPTG, but without induction of the plasmid-borne ftsZ and mreBCD genes. These plates gave viable counts reflecting the effect of the mreB/ftsZ-mutant combinations in the liquid media. CFU measurements are an average of three independent repeats (n=3). Note that the expression of MreBD285A reduced CFU as cells elongate (compare CFU/ml when MreBwt or MreBD285A is overexpressed and the FtsZwt allele is used for VIP205 complementation). Using FtsZP203Q for VIP205 complementation in cells overexpressing MreBwt reduced cell viability four-fold (see corrected fold complementation). In contrast, FtsZP203Q expression in VIP205 cells also expressing MreBD285A yielded a near 200-fold higher CFU (in growing cultures). Images of serially diluted plates are shown in Supplementary Figure S5C along with plate images showing MreBD285A complementation for all the FtsZ variants identified in this study.