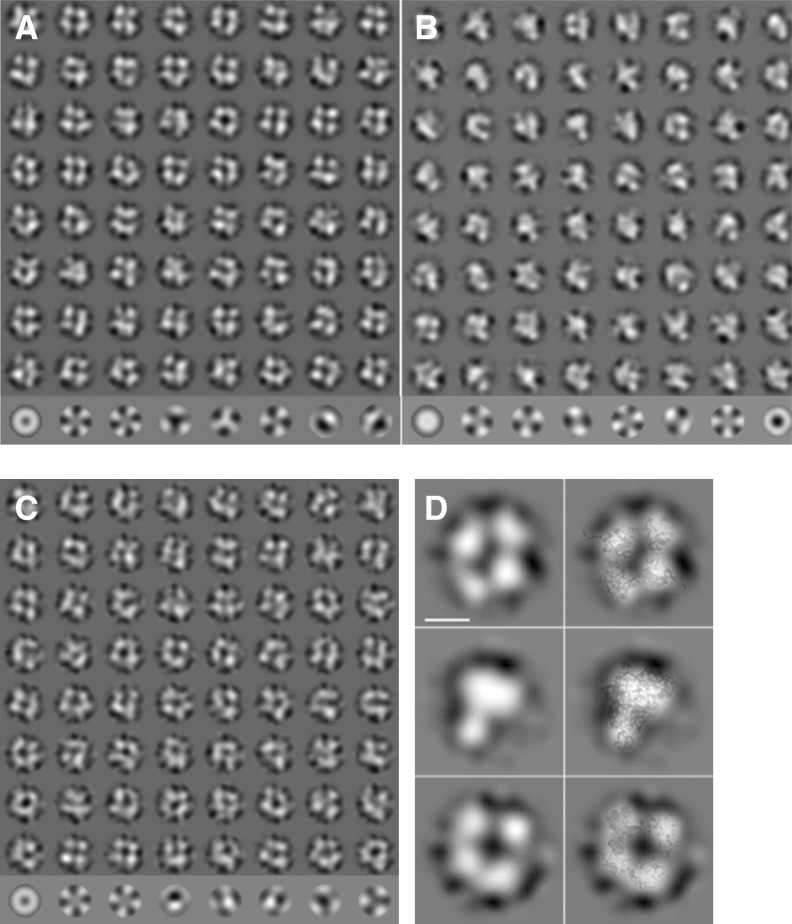
FIGURE 3.
Electron microscopy of negatively stained SBV N protein preparations. (A) Class averages (rows 1–8) and eigenimages (row 9) of SBV N native particles. Most of the class averages are consistent with a tetramer. However, there are also a few other oligomeric states such as trimer (row 8, column 3) or pentamer (row 8, column 6). (B) Class averages of refolded SBV N particles (rows 1–8) and eigenimages (row 9). The class averages show a heterogeneous particle population with trimers (e.g., row 1, column 1; row 2, column 8; row 3, column 6; row 7, column 1) as major component and some tetramers (e.g., row 1, column 4; row 1, column 6). (C) Class averages of the refolded SBV N-RNA complex (rows 1–8) and eigenimages (row 9). Most class averages show a tetramer. However, some class averages are consistent with other stoichiometries. (D) Selected class averages (left) and class average with the respective crystal structures overlaid (right). (Top) native SBV N; (middle) refolded SBV N; (bottom) refolded SBV N RNA complex. The overlay shows a good match between the crystal structures and the electron microscopic class averages. The length of the scale bar equals 5 nm.