Abstract
Natural Killer T (NKT) cells are a subset of T lymphocytes that recognize a wide variety of lipid antigens presented by CD1 molecules. NKT cells exhibit rapid activation after recognition of cognate antigens, secrete abundant amounts of T helper (Th) 1, Th2, and Th17 cytokines within hours of activation and shape the immune response through subsequent activation of dendritic, NK, T and B cells. NKT cells therefore play central roles in antimicrobial and anticancer immunity and in modulation of various autoimmune disorders. Consequently, recent research has focused on the discovery of microbial and self-antigens involved in NKT cell activation. In this chapter, we discuss different strategies for studying antigen recognition by NKT cells including CD1d tetramer-based approaches and in vitro assays characterizing NKT cell activation in response to lipid antigen presentation. While toll-like receptor (TLR) agonists and cytokines such as IL-12 are critical for NKT cell activation in vivo, particularly in the context of microbial infection, methods for detection of TLR- and cytokine-dependent NKT cell activation will not be discussed in this section.
Keywords: CD1d, Natural Killer T cells, Lipid antigen presentation, Tetramer, α-Galactosylceramide
1. Introduction
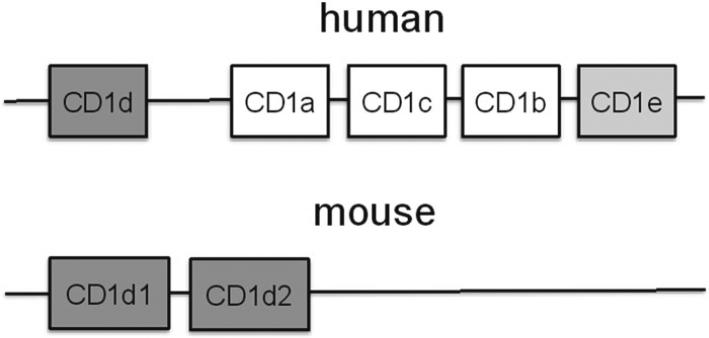
The CD1 gene family consists of five members in humans and two highly related members in mouse (1, 2). In humans, these consist of type 1 (CD1a–c) and type 2 (CD1d) that are expressed on the cell surface and which differentially traffic through the endolysomal system for sampling the lipid milieu within specific cellular locales (1, 2). The fifth human CD1 member, CD1e, is not expressed on the cell surface and plays a role in the regulation of the other CD1 family members (3). In mouse, only type 2 CD1 proteins are expressed and include two highly related CD1d molecules, CD1d1 and CD1d2 (for genomic organization please refer to Fig. 1) (4). CD1d1 and CD1d2 are highly homologous and in certain strains of mice such as C57Bl/6 the CD1d2 gene product is nonfunctional due to a loss-of-function mutation in its open reading frame (4).
Fig. 1.
Genomic organization of the human and murine CD1 locus.
All of the CD1 proteins are characteristically involved in presenting endogenous (host) and exogenous (microbial) lipid antigens to distinct subsets of T cells (1, 2). There are two types of T cells which are involved in recognizing lipid antigens in the context of CD1 on an antigen presenting cell (APC). The T cells which recognize CD1d in mouse and human are considered as natural killer T cells (NKT cells) in view of their expression of both NK (NK1.1) and T cell (e.g., T cell receptor) markers (5, 6). Further, NKT cells include those that express an invariant T cell receptor (TCR) alpha chain in association with a variety of TCR-β chains (Table 1). These so-called invariant (i) NKT cells are the variety that are currently accessible to careful interrogation since they recognize an antigen, α-galactosylceramide (α-GalCer), in the context of CD1d (18). A second class of NKT cells exist which express a variable, but restricted, array of TCR-α and TCR-β chains and are considered to be noninvariant (ni) NKT cells (Table 1) (8, 9). Although these niNKT cells are restricted by CD1d and certain antigens have been defined as determinant of recognition in the context of CD1d (e.g., sulfatides), these are less amenable to study as will be discussed further below given the relative range of antigens that have been identified to date which are responsible for their recognition and activation. We will not discuss cells that are restricted by human CD1a–c which also express a variety of TCR-α and TCR-β chains and which are responsible for recognizing a variety of poorly characterized endogenous antigens as well as a wide variety of exogenous microbial antigens from mycobacterial and non-mycobacterial sources (1). With the recent development of type 1 CD1 tetramers, the comments below will likely be extensible to these subsets of T cells as well (10).
Table 1.
Subtypes of NKT cells
| NKT cell | T cell receptor (TCR) | Detection/deletion |
|---|---|---|
| Invariant (Type I) | Vα14Jα18 in mice | Detected by αGalCer/CD1d tetramers |
| Vα24Jα18 in humans | Deleted in Jα18-/- mice (TCR Jα18 deletion) | |
| Biased Vβ repertoire | Deleted in CD1d-/- mice (impaired thymic selection) | |
| Noninvariant (Type II) | Oligoclonal TCR repertoire | Subgroup detected by sulfatide/CD1d tetramers |
| Not deleted in Jα18-/- mice | ||
| Deleted in CD1d-/- mice (impaired thymic selection) | ||
CD1d is synthesized as a non-covalent heterodimer with β2-microglobulin (β2m) in the endoplasmic reticulum (ER), where it is loaded and presumably stabilized by endogenous lipid antigens (1, 11). CD1d then traffics to the cell surface and presents lipids acquired in the secretory pathway through the actions of microsomal triglyceride transfer protein (MTP), an ER resident lipid transfer protein (1, 12–15). Subsequently, CD1d undergoes endocytosis and reaches endosomes (murine CD1d (mCD1d) and human (hCD1d)) and lysosomes (mCD1d only) where lipid transfer and editing molecules assist in the exchange of bound antigens against other endogenous and exogenous lipids (1). CD1d thus surveys the cell for lipid antigens and presents a wide variety of self and foreign lipids acquired in diverse subcellular compartments (1).
Methods for characterization of antigen recognition by NKT cells are based on CD1d and rely either on direct detection of CD1d–lipid complexes bound to the NKT cell receptor or on measurement of NKT cell activation and cytokine secretion in response to CD1d-restricted lipid antigen presentation (16). Direct detection of antigen recognition by NKT cells is based on high-affinity multimers, usually tetramers, consisting of streptavidin-bound CD1d/β2m complexes loaded with the lipid of interest. Conjugation of fluorophores to streptavidin allows detection of CD1d–lipid complex binding to the NKT cell receptor by flow cytometry. Using this approach, CD1d tetramers loaded with the marine sponge glycosphingolipid α-galactosylceramide (αGalCer) or its analogues (e.g., PBS57) allow sensitive and specific detection of murine and human iNKT cells, a subgroup of NKT cells expressing a semi-invariant TCR consisting of an invariant TCR alpha chain (Vα14Jα18 (17) in mice, Vα24Jα18 in humans) paired with a restricted set of TCR-β chains (Table 1 and Fig. 2) (18). In addition, sulfatide-loaded CD1d tetramers have been shown to allow detection of a population of niNKT cells, a subgroup of NKT cells expressing a more diverse though still biased TCR repertoire as discussed above (19, 20). While CD1d tetramers allow for unambiguous direct detection of NKT cell antigen recognition, NKT cell-activating CD1d-restricted lipids can fail in tetramer-based approaches due to the inability to form stable complexes with CD1d (e.g., lysophospholipids) or low affinity to the T cell receptor (e.g., microbial-derived diacylglycerols).
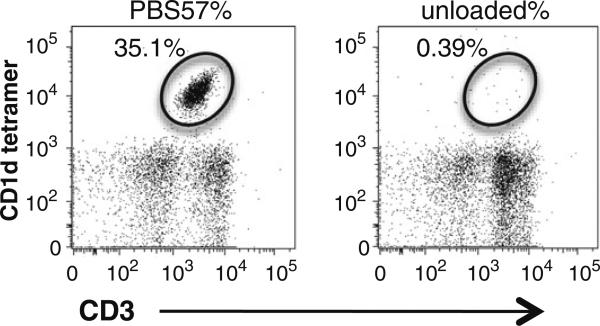
Fig. 2.
Flow cytometry staining of iNKT cells. Invariant NKT cells are shown as PBS57/CD1d tetramer-positive, CD3intermediate T cells (left panel). Unloaded CD1d tetramers served as control (right panel).
In cases where potential antigens fail in generation of tetramers, antigen recognition by NKT cells can be demonstrated by upregulation of activation markers or secretion of cytokines by NKT cells in response to CD1d-restricted presentation of these lipids. To this end, CD1d-expressing antigen presenting cells (APC) are loaded with the respective lipids and cocultured with NKT cells in vitro or, alternatively, injected into mice for analysis of antigen presentation in vivo. Many lipids will in addition to functioning as antigens act as bioactive molecules that induce lipid signaling, metabolic alterations, and changes in lipid raft structure; effects that can be largely prevented in cell-free APC assays. In this approach, recombinant CD1d is coated onto plates, loaded with the lipids of interest and subsequently tested for presentation of these lipids to NKT cells.
In the following sections, we will discuss the generation of custom-made CD1d tetramers, isolation of NKT cells, and detection of NKT cells by tetramers. We will also outline strategies for characterization of antigen recognition by NKT cells based on their activation upon CD1d-restricted presentation of lipid antigens.
2. Materials
2.1. Consumables and Lab Equipment
Donor mice.
Borosilicate glass tubes (e.g., Pyrex ®).
Syringes, 25 G needles.
15 and 50 mL Conical tubes and Flow cytometry tubes.
150 mm Non-plugged Pasteur pipettes.
Nylon mesh (e.g., BD Falcon cell strainers 40 and 70 μm).
96-Well flat bottom plates.
Flow cytometer with possibility of cell sorting.
2.2. Generation of CD1d–Lipid Tetramers
PE- or APC-conjugated preloaded murine and human CD1d tetramers (NIH Tetramer Core Facility or different commercial vendors) or custom-made CD1d tetramers (see Subheading 3.1). For instructions on NIH tetramer orders please refer to http://tetramer.yerkes.emory.edu.
Biotinylated mouse or human CD1d monomer (NIH Tetramer Core Facility).
Streptavidin-PE or Streptavidin-APC.
Synthetic or (ideally HPLC-) purified lipids (various sources, e.g., Avanti Polar Lipids, Inc.).
2.3. Cell Lines
Primary APCs; CD1d-transfected and untransfected cell lines.
Primary NKT cells or NKT cell lines/clones/hybridomas.
Tissue culture medium: DMEM or RPMI-1640 containing 10% fetal bovine serum and 2 mM l-glutamine.
Percoll Plus.
2.4. Buffers
1× PBS sterile.
ACK lysis buffer: 0.15 M ammonium chloride (8.29 g NH4Cl), 10 mM potassium bicarbonate (1 g KHCO3) and 0.1 mM Na2EDTA (37.2 mg), add 800 mL H2O and adjust pH to 7.2–7.4 with 1 N HCl. Finally, add H2O to 1 L and filter through a 0.2-μm filter to sterilize and store at room temperature.
Staining buffer: 1× PBS, 1% BSA.
2.5. Antibodies and Reagents
6B11 Anti-human iNKT TCR antibody (e.g., eBioscience).
Directly conjugated antibodies of choice for surface and intra-cellular staining of NKT cells.
Unlabeled streptavidin.
Cytofix/Cytoperm Plus kit (BD Biosciences).
Reagents for detection of NKT cell activation such as ELISA kits or flow cytometry antibodies.
3. Methods
3.1. Generation of CD1d–Lipid Tetramers
Tetramers of CD1d allow high-affinity single-cell detection of T cells that respond to CD1d-restricted lipid antigens. Strategies rely on loading of CD1d with the respective lipids, followed by streptavidin-mediated tetramerization of biotinylated CD1d/β2m monomers and flow cytometry-based detection of CD1d/lipid complexes bound to the NKT cell receptor. Here, we describe the generation of lipid-loaded CD1d tetramers. For ready-to-use CD1d tetramers, refer to Note 1.
For each staining between 0.1 and 4 μg CD1d tetramer are required and are loaded with a 40-fold molar excess of the lipid of interest. The amount of lipid required can be calculated as follows:
mCD1d = Microgram of biotinylated CD1d (0.1–4 μg/staining)
ME = Molar excess of lipid antigen (e.g., 40-fold)
MwAntigen = Molecular weight of antigen
MwCD1d = Molecular weight of biotinylated CD1d (48,579 Da)
For example, for the generation of 4 μg of CD1d loaded with α-galactosylceramide (α-GalCer), the following calculation would apply
If lipids are resuspended in nonpolar solvents, transfer the required amount of lipid to a sonication-resistant borosilicate glass tube and dry the lipid under nitrogen stream. Add 1× PBS to obtain a 0.1–2 mg/mL solution of the lipid (see Note 2).
Mix 1–4 μg of biotinylated CD1d with the required amount of lipid and incubate at room temperature for 12–16 h (see Note 3).
For tetramerization, add aliquots of 1 μL (500 ng) of streptavidin–PE or streptavidin–APC every 10 min for a total of ten times. Mix well after each addition (see Note 4).
In case of stable complexes such as α-galactosylceramide/CD1d, tetramers can be stored at 4°C for weeks. However, in other cases, such as sulfatide/CD1d complexes, tetramers should be used immediately.
3.2. Extraction of Mononuclear Cells for NKT Cell Analysis
A general protocol for extraction of mononuclear cells from liver, spleen, and thymus is outlined. For an overview of expected cell numbers, see Note 5.
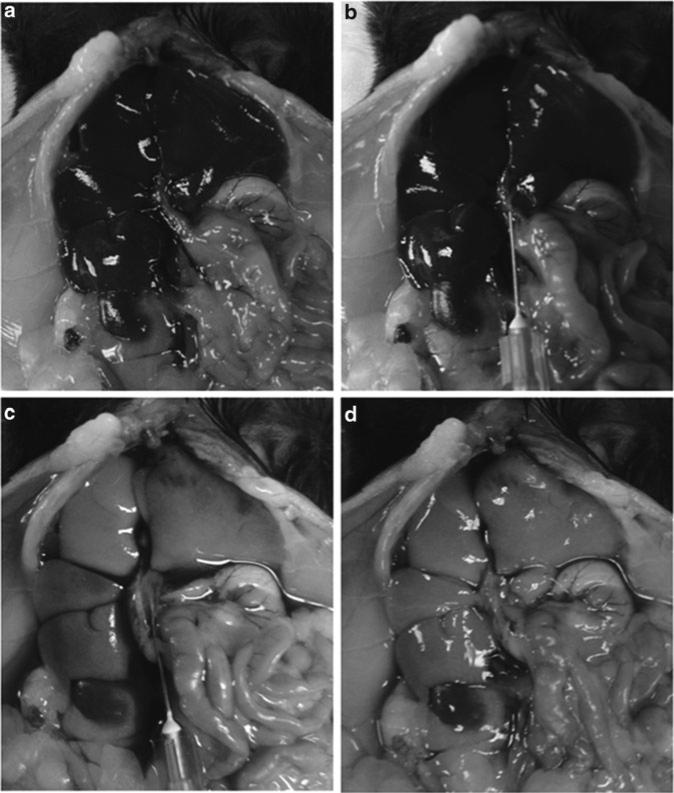
Immediately after euthanization of a mouse, open the peritoneal cavity and perfuse the liver with 10 mL of 1× PBS through the portal vein using a 25 G needle. After starting perfusion the inferior vena cava should be cut to facilitate flow. This step will remove blood cells but maintain liver mononuclear cells. Immediately upon perfusion, the liver should turn pale as a sign of successful perfusion (Fig. 3). Otherwise relocate the needle to make sure it is placed in the portal vein. The thymus and spleen are dissected out and all tissues are placed in a separate tube with cold PBS.
Harvest the liver and press gently through a nylon mesh (70 μm for liver, 40 μm for thymus and spleen) using the back of a 3 mL syringe plunger. Collect the filtered solution, transfer to a 50 mL conical tube, add 1× PBS for a total of 50 mL, and centrifuge at 400 × g for 5 min at 4°C.
- For the liver preparation only.
- Carefully aspirate the supernatant, fill the tube with 50 mL of 1× PBS, and centrifuge at 400 × g for 5 min at 4°C.
- Aspirate supernatant and resuspend the pellet in 4 mL of 40% Percoll. Transfer to a conical 15 mL tube.
- Carefully underlay 2 mL of 60% Percoll ensuring that 40% and 60% Percoll are not mixed (see Note 6).
- Centrifuge tubes at 700 × g for 20 min at 4°C. Important : Make sure the brake of the centrifuge is switched off since this will otherwise lead to a mixture of the two Percoll layers.
- After centrifugation, hepatocytes are on top and liver mononuclear cells are at the interphase of the Percoll gradient (at the 2 mL level).
- Remove hepatocytes from top by careful aspiration avoiding inadvertent aspiration of the interphase or a mixture of both layers.
- Using a 5 mL pipette, collect LMNCs by aspiration at the interphase. Transfer cells to a 50 mL tube, add 1× PBS for a total of 50 mL, and centrifuge at 400 × g for 5 min at 4°C.
Aspirate the supernatant and resuspend pelleted cells in ACK lysis buffer for lysis of red blood cells (not required for thymus). Incubate cells for 5 min at room temperature, then add 45 mL of 1× PBS and centrifuge at 400 × g for 5 min at 4°C. Repeat washing step with another 50 mL of 1× PBS.
Resuspend cells in staining buffer.
Fig. 3.
Perfusion of a mouse liver. (a) Situation before perfusion with portal (right) and inferior cava (left) vein entering the liver. (b) Insertion of a needle into the portal vein. (c) Start of perfusion of the liver. (d) Situation after perfusion of the liver. Please note the change in liver color after perfusion as a sign of successful removal of blood cells.
3.3. Tetramer and Antibody Staining of iNKT Cells
A protocol for flow cytometry-based detection of iNKT cells is outlined. For an overview of methods for NKT detection, please see Note 7. For an high-throughput adaptation, please see Note 8).
Pellet mononuclear cells by centrifugation at 400 × g for 5 min at 4°C.
Carefully aspirate supernatant and add CD1d–lipid tetramer in 50 μL of staining buffer. Incubate for 30 min at 4°C.
Without washing, add fluorochrome-conjugated antibodies (see Notes 9 and 10) for surface staining at pre-tritrated concentrations (usually 0.1–5 μg/mL) in 50 μL of staining buffer. Incubate for an additional 30 min at 4°C.
For washing, add 2 mL of staining buffer and centrifuge at 400 × g for 5 min at 4°C. Carefully aspirate supernatant.
For direct flow cytometry analysis, resuspend cells in staining buffer and analyze.
- For analysis of intracellular molecules such as transcriptional regulators or cytokines (see Note 11).
- Resuspend cells in 250 μL Cytofix/Cytoperm buffer for 20 min at 4°C for fixation and permeabilization.
- Wash cells by the addition of 2 mL of 1 × Perm/Wash buffer and centrifugation at 600 × g for 5 min at 4°C.
- Add pre-titrated directly conjugated antibodies (usually 0.1–5 μg/mL) for intracellular staining in 100 μL of 1 × Perm/Wash buffer and incubate for 30 min at 4°C.
- Wash cells by addition of 2 mL of 1 × Perm/Wash buffer and centrifugation at 600 × g for 5 min at 4°C.
- Resuspend cells in staining buffer for flow cytometry analysis.
3.4. Characterization of Functional NKT Cell Responses to Lipid Antigens Using a Coculture Approach
Transfer APCs (for choice of APCs please see Note 12) in the appropriate cell culture medium to a suitable tube and add lipids of interest aiming for a series of tenfold dilutions with final lipid concentrations of 10 μg/mL to 1 ng/mL (see Note 13).
Incubate for 4–16 h at 37°C in a tissue culture incubator (see Note 14).
Count APCs and add 2 × 104 to 1 × 105 APCs per well in 100 μl of the appropriate cell culture medium to 96-well flat bottom plates (see Notes 15 and 16). Use triplicates for each condition.
Remove unbound lipid by 3–5 washing steps using the appropriate tissue culture medium. In case of non-adherent APCs, centrifugation at 400 × g for 5 min is required for washing (see Note 17).
After the last washing step, aspirate supernatant and add 100 μL of the appropriate tissue culture medium.
Resuspend NKT cells in the same tissue culture medium used for the APCs and add 2 × 105 primary iNKT cells or 5 × 104 cells from an iNKT cell clone or hybridoma in 100 μL (see Note 18).
Incubate at 37°C in a tissue culture incubator (see Note 19).
Analyze NKT cell activation by ELISA- or flow cytometry-based detection of cytokine secretion or flow cytometry-based detection of upregulation of NKT activation markers (see Note 20).
3.5. Characterization of Functional NKT Cell Responses to Lipid Antigens Using an APC-Free Assay
APC-free assays for lipid antigen presentation are based on CD1d-β2-monomers that are bound to tissue culture plates and allow for loading of lipids and presentation to NKT cells.
Load 0.25 μg of CD1d in 100 μL 1× PBS per well onto 96-well flat bottom tissue culture plates. Use triplicates for each experimental condition.
Incubate for 16 h at 37°C in a humidified incubator to allow for binding of CD1d to the plate (see Note 21).
Wash three times with 200 μL of 1× PBS to remove unbound CD1d.
Add lipids in triplicates in a series of tenfold dilutions in 100 μL of 1× PBS per well as described in Subheading 3.4, step 1.
Incubate for 16 h at 37°C in a humidified incubator to allow for spontaneous lipid loading onto CD1d (see Note 22).
Wash five times with 200 μL of 1× PBS to remove unbound lipid.
Add 2 × 105 primary iNKT cells or 5 × 104 cells from an iNKT cell clone or hybridoma in 200 μL of tissue culture medium.
Analyze NKT cell activation as described in Subheading 3.4, step 8.
Acknowledgment
The work was supported by NIH grants DK51362, DK44319, DK53056, DK88199, the Harvard Digestive Diseases Center (DK034856) (to R.S.B.); the Deutsche Forschungsgemeinschaft (Ze 814/1-1, Ze 814/4-1), and the FP7-PEOPLE program of the European Commission (Marie Curie International Reintegration Grant 256363) (to S.Z.); the Deutsche Forschungsgemeinschaft (OL 324/1-1) (to T.O.) E.M. was financed through the Norwegian PSC research center, Caroline Musæus Aarsvolds fund, and the Unger - Vetlesen Medical Fund.
Footnotes
Fluorescently labeled CD1d tetramers loaded with α-galactosylceramide (α-GalCer) analogues are available from the National Institutes of Health (NIH) Tetramer Core Facility (http://tetramer.yerkes.emory.edu/available/nonclassical_cd1d) and other commercial sources and allow for sensitive and specific detection of iNKT cells.
Depending on the solubility of the lipid of interest, detergents such as Tween or Triton-X might have to be added in low concentrations and the aqueous solution might have to be sonicated for 10 min in a water bath at room temperature or 37°C to ensure that lipids dissolve in PBS. However, addition of detergents might interfere with subsequent loading of lipids onto CD1d and prevent the formation of stable CD1d/lipid complexes.
It is noteworthy that recombinant CD1d, probably independent of the choice of expression system, is likely to contain endogenous lipids bound to the CD1d groove. Loading of CD1d with exogenous lipids will therefore rely on their ability to replace the endogenous lipid(s) bound to “unloaded” CD1d.
Phycoerythrin (PE) and Allophycocyanin (APC) are recommended as fluorophores since they exhibit strong fluorescence allowing detection of binding of low affinity antigen tetramers. Use of the respective fluorophores will also depend on the configuration of the flow cytometer.
Tissues vary significantly in the relative and absolute numbers of iNKT cells (18). However, little is known about the distribution of niNKT cells. The relative abundance of iNKT cells is highest in murine livers with up to 30–40% of T cells representing iNKT cells based upon analysis with αGalCer-loaded CD1d tetramers (18). In contrast, other human and murine lymphoid and non-lymphoid tissues contain low relative numbers of iNKT cells, usually less than two percent of T cells. Still, due to differences in the total T cell count, absolute numbers of iNKT cells are higher in spleen, thymus, and intestine compared to the liver. Thus, a murine liver yields about 1–2 × 106 mononuclear cells, with about 0.25–0.5 × 106 iNKT cells assuming 60% T cells and 40% of T cells expressing the iNKT TCR. From spleen, about 1 × 106 iNKT cells can be extracted assuming 1 × 108 total cells, 50% T cells, and 2% of T cells expressing the iNKT TCR.
The easiest way to achieve this is to put a Pasteur pipette at the bottom of the tube and to fill it with 2 mL of 60% Percoll. The Pasteur pipette will ensure slow release of Percoll and avoid a mixture of the two layers. In case the Pasteur pipette does not release the Percoll, carefully lift it so that it does not touch the bottom of the tube.
Invariant NKT cells can be detected in a sensitive and highly specific manner by CD1d tetramers loaded with the marine sponge glycosphingolipid α-GalCer (18) or its analogue PBS-57. Such tetramers are available in preloaded, fluorophore-conjugated formats from the NIH Tetramer Core Facility and different commercial vendors. Consistent with cross-species reactivity of CD1d, murine α-GalCer/CD1d tetramers can be used for staining of human iNKT cells (15). In addition to α-GalCer/CD1d tetramers, human but not mouse iNKT cells can be detected with a monoclonal antibody directed against a unique determinant in the complementarity determining region (CDR) 3 region of the human invariant (Vα24-Jα18) TCR chain (21) or by direct staining of the human iNKT TCR using antibodies against TCR Vα24 and TCR Vβ 11. Furthermore, CD1d can be loaded with lipids of interest for tetramer-based characterization of antigen recognition by iNKT cells as described above.
We usually stain cells in 5 mL flow cytometry tubes. However, in case of multiple stainings, cells can also be stained in 96-well round or V-bottom plates. In this case, washing steps only allow lower volumes of washing buffer (200 μl). Accordingly, two instead of one washing cycles should be performed at each step.
- Molecules such as CD4 (mouse and human) and CD8 (human) to distinguish CD4– CD8– double negative from CD4+ and CD8+ iNKT cells.
- NK cell markers typically expressed by NKT cells such as CD161 (human), NK1.1 (selected inbred mouse strains including C57BL/6, FVB/N, and NZB, but not AKR, BALB/c, CBA/J, C3H, DBA/1, DBA/2, NOD, SJL, and 129) and CD49b (most inbred mouse strains including BALB/c, C57BL/6, C3H, CBA, DBA, AKR, SJL, and 129).
- Differentiation (NK1.1, CD44, CD122), activation (CD69, CD38, CD25, CD95), effector memory markers (CD45ROhi, CD62Llo) expressed by NKT cells (21, 22). It is noteworthy that some of these markers, such as CD69, are acquired during thymic development of NKT cells and are largely independent of interaction with CD1d following thymic emigration thus serving as a marker for central NKT development and maturation (23). In contrast, other markers, such as NK1.1 are at least partially acquired in the periphery after thymic emigration, are dependent on CD1d-restricted antigen presentation in peripheral lymphoid and non-lymphoid organs and thus serve as markers for post-thymic NKT maturation and homeostasis (23). For additional information, please refer to excellent reviews on this topic (22, 24, 25).
- Dead cell markers such as 7-AAD, propidium iodide or 4-,6-Diamidin-2-phenylindol (DAPI).
In addition to αGC-loaded CD1d tetramers, human iNKT cells can be identified by the 6B11 antibody that recognizes a unique determinant in the complementarity determining region (CDR) 3 region of the human invariant (Vα24-Jα18) TCR chain (21). Alternatively, iNKT cells can be detected by direct staining of the human iNKT TCR using antibodies against TCR Vα24 and TCR Vβ 11 but this approach is less specific compared to αGC/CD1d-tetramer- and 6B11-staining (26). In contrast to human iNKT cells, detection of mouse iNKT cells is dependent on αGC/CD1d-tetramers since iNKT TCR antibodies are not commercially available.
For intracellular staining, transcriptional regulators such as the largely NKT cell-specific promyelocytic leukemia zinc finger (PLZF) (27, 28), the IL-17-lineage marker RORγt (7), the regulatory marker FoxP3 (29), and many other transcription factors expressed by NKT cells (24) should be considered. In addition, cytokines produced by NKT cells such as IFN-γ, IL-4, IL-10, IL-13, and IL-17 can be detected by intracellular cytokine staining, particularly after activation of NKT cells in vitro or in vivo.
For coculture systems, the choice of APC is most relevant. When synthetic or purified exogenous lipid antigens are used, the monomorphic nature of CD1d allows the use of many different types of APCs including primary CD1d-expressing APCs such as DCs, monocytes, and B cells; CD1d transfectants such as murine fibroblasts (L-CD1d), murine T cell lymphoma (RMAS-CD1d), murine B cell lymphoma (A20-CD1d), and human CD1d-transfected C1R B cells, 293 human embryonic kidney cells, and HeLa cervical cancer cells. While CD1d-transfected cell lines are readily available and easy to handle, lipids that require processing for loading onto CD1d might be presented more efficiently by primary APCs that are capable of extensive endolysosomal processing such as DCs.
In case hydrophobicity prevents direct addition of lipids to aqueous solutions, proceed as described in step 1 of Subheading 3.1, transfer lipids from nonpolar solvents to a sonication-resistant borosilicate glass tube, dry lipids under nitrogen stream, and dissolve in tissue culture medium at the final concentration by sonication in a water bath.
CD1d lipids that load at the cell surface require less time for efficient transfer onto CD1d than lipids dependent on endolysosomal loading (30). Similarly, processing-dependent antigens require longer incubation times than non-processing-dependent lipids.
It is critical to include a CD1d-negative control to demonstrate CD1d-restriction of NKT cell activation. In case of CD1d-transfected cell lines, mock-transfected cells serve as control. In case of primary APCs, antibody-mediated CD1d blocking is the method of choice. To this end, block CD1d by the addition of monoclonal, unlabeled, azide-free antibodies between steps 5 and 6. An antibody concentration of 10 μg/mL is usually sufficient. Clones 19 G11 for murine CD1d and 51.1 for human CD1d block CD1d-restricted antigen presentation of a variety of different APCs (15, 31). It should be noted, however, that antibody blocking of CD1d is cell-type specific despite the monomorphic structure of CD1d (31). Notable exceptions of monoclonal antibodies with universal CD1d blocking across different cell types, such as 19 G11, exist (31).
The optimal ratio of APCs to primary iNKT cells and iNKT cell clones/hybridomas varies depending on the kind of APC and NKT cell used for coculture experiments. Pilot studies should be performed with different relative and absolute amounts of APCs and NKT cells.
Extensive washing is preferred in order to ensure that all unloaded lipid antigen is removed in order to avoid processing and presentation by CD1d bearing T cells when analyzing murine T cells. Human CD1d is not typically expressed on T cells so is usually not an issue when analyzing human systems (32).
Primary NKT cells, NKT cell clones and lines, and NKT cell hybridomas can be used. Primary NKT cells are difficult to extract in sufficient quantities but allow for analysis of polyclonal NKT cells producing of a wide variety of NKT cell cytokines. In contrast, NKT cell clones and particularly hybridomas can be easily expanded and maintained in culture but are restricted in the diversity of secreted cytokines (in hybridomas usually restricted to IL-2). In addition, it is important to note that iNKT cells express a semi-invariant TCR with various possible TCR-β chains that affect antigen recognition. Primary NKT cells thus offer a source of oligoclonal NKT cells with various TCR-β chains, while iNKT cell clones and hybridomas are monoclonal and limited to one particular TCR-β chain.
The required time for coculture is dependent on the type of assay used for the detection of NKT cell activation. Intracellular cytokines and upregulation of activation markers such as CD69 can often be detected after 12–24 h of stimulation by flow cytometry. In addition, secretion of IL-4, as detected by ELISA, reaches its peak at 24 h, while IFN-γ peaks at 72 h of coculture. For ELISpot assays, 16–24 h of coculture are sufficient.
Primary NKT cells usually secrete abundant amounts of IFN-γ and IL-4 that can be detected in ELISA- and flow cytometry-based approaches. NKT cell clones and particularly hybridomas often lose the ability to secrete IFN-γ and IL-4. In these cases, IL-2 is the preferred cytokine for detection of activation. As an activation marker, CD69 shows upregulation of NKT cell surface expression within hours of activation and can be used for indirect detection of lipid antigen recognition by NKT cells.
Alternative approaches for enhanced CD1d binding such as streptavidin-coated plates or the use of CD1d-Fc fusion proteins on protein G-coated plates exist but are usually not necessary for activation of NKT cells by plate bound CD1d.
The addition of purified lipid transfer proteins such as microsomal triglyceride transfer protein (commercially available from different vendors) might facilitate the transfer of lipids onto CD1d (13).
References
- 1.Cohen NR, Garg S, Brenner MB. Antigen presentation by CD1 lipids, T cells, and NKT cells in microbial immunity. Adv Immunol. 2009;102:1–94. doi: 10.1016/S0065-2776(09)01201-2. [DOI] [PubMed] [Google Scholar]
- 2.Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929–941. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- 3.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 4.Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- 5.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L. NKT cells: what's in a name? Nat Rev Immunol. 2004;4:231–237. doi: 10.1038/nri1309. [DOI] [PubMed] [Google Scholar]
- 6.Zeissig S, Kaser A, Dougan SK, Nieuwenhuis EE, Blumberg RS. Role of NKT cells in the digestive system. III. Role of NKT cells in intestinal immunity. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1101–G1105. doi: 10.1152/ajpgi.00342.2007. [DOI] [PubMed] [Google Scholar]
- 7.Michel ML, et al. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardell S, et al. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu YH, et al. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasmar AG, et al. CD1b tetramers bind alpha}{beta T cell receptors to identify a myco-bacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208(9):1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odyniec AN, et al. Regulation of CD1 antigen-presenting complex stability. J Biol Chem. 2010;285:11937–11947. doi: 10.1074/jbc.M109.077933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dougan SK, Rava P, Hussain MM, Blumberg RS. MTP regulated by an alternate promoter is essential for NKT cell development. J Exp Med. 2007;204:533–545. doi: 10.1084/jem.20062006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dougan SK, et al. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaser A, et al. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur J Immunol. 2008;38:2351–2359. doi: 10.1002/eji.200738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeissig S, et al. Primary deficiency of microsomal triglyceride transfer protein in human abetalipoproteinemia is associated with loss of CD1 function. J Clin Invest. 2010;120:2889–2899. doi: 10.1172/JCI42703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tupin E, Kronenberg M. Activation of natural killer T cells by glycolipids. Methods Enzymol. 2006;417:185–201. doi: 10.1016/S0076-6879(06)17014-7. [DOI] [PubMed] [Google Scholar]
- 17.Koseki H, et al. Homogenous junctional sequence of the V14+ T-cell antigen receptor alpha chain expanded in unprimed mice. Proc Natl Acad Sci U S A. 1990;87:5248–5252. doi: 10.1073/pnas.87.14.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda JL, et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–754. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halder RC, Aguilera C, Maricic I, Kumar V. Type II NKT cell-mediated anergy induction in type I NKT cells prevents inflammatory liver disease. J Clin Invest. 2007;117:2302–2312. doi: 10.1172/JCI31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jahng A, et al. Prevention of autoimmunity by targeting a distinct, noninvariant CD1d-reactive T cell population reactive to sulfatide. J Exp Med. 2004;199:947–957. doi: 10.1084/jem.20031389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montoya CJ, et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology. 2007;122:1–14. doi: 10.1111/j.1365-2567.2007.02647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 23.McNab FW, et al. The influence of CD1d in postselection NKT cell maturation and homeostasis. J Immunol. 2005;175:3762–3768. doi: 10.4049/jimmunol.175.6.3762. [DOI] [PubMed] [Google Scholar]
- 24.Das R, Sant'Angelo DB, Sant'Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 26.Lee PT, et al. Testing the NKT cell hypothesis of human IDDM pathogenesis. J Clin Invest. 2002;110:793–800. doi: 10.1172/JCI15832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteiro M, et al. Identification of regulatory Foxp3+ invariant NKT cells induced by TGF-beta. J Immunol. 2010;185:2157–2163. doi: 10.4049/jimmunol.1000359. [DOI] [PubMed] [Google Scholar]
- 30.Yu KO, et al. Production and characterization of monoclonal antibodies against complexes of the NKT cell ligand alpha-galactosylceramide bound to mouse CD1d. J Immunol Methods. 2007;323:11–23. doi: 10.1016/j.jim.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roark JH, et al. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–3127. [PubMed] [Google Scholar]
- 32.Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]