The position of the spindle apparatus determines the plane of cell cleavage and, therefore, the size and position of daughter cells, as well as the decision between symmetric and asymmetric cell division. We show that asymmetry in cortical actin and, remarkably, an endoplasmic reticulum–localized protein contribute to proper spindle positioning in the Caenorhabditis elegans embryo.
Abstract
The microtubule spindle apparatus dictates the plane of cell cleavage in animal cells. During development, dividing cells control the position of the spindle to determine the size, location, and fate of daughter cells. Spindle positioning depends on pulling forces that act between the cell periphery and astral microtubules. This involves dynein recruitment to the cell cortex by a heterotrimeric G-protein α subunit in complex with a TPR-GoLoco motif protein (GPR-1/2, Pins, LGN) and coiled-coil protein (LIN-5, Mud, NuMA). In this study, we searched for additional factors that contribute to spindle positioning in the one-cell Caenorhabditis elegans embryo. We show that cortical actin is not needed for Gα–GPR–LIN-5 localization and pulling force generation. Instead, actin accumulation in the anterior actually reduces pulling forces, possibly by increasing cortical rigidity. Examining membrane-associated proteins that copurified with GOA-1 Gα, we found that the transmembrane and coiled-coil domain protein 1 (TCC-1) contributes to proper spindle movements. TCC-1 localizes to the endoplasmic reticulum membrane and interacts with UNC-116 kinesin-1 heavy chain in yeast two-hybrid assays. RNA interference of tcc-1 and unc-116 causes similar defects in meiotic spindle positioning, supporting the concept of TCC-1 acting with kinesin-1 in vivo. These results emphasize the contribution of membrane-associated and cortical proteins other than Gα–GPR–LIN-5 in balancing the pulling forces that position the spindle during asymmetric cell division.
INTRODUCTION
During the development of higher eukaryotes, elaborate regulatory pathways control the timing, location, and orientation of cell division. Most cell divisions create daughter cells of equal size and fate, thereby supporting exponential increases in cell numbers. In development, however, certain asymmetric cell divisions generate unequal daughter cells, which creates cell diversity and helps maintain tissue-specific stem cells (Galli and van den Heuvel, 2008; Knoblich, 2008). Such divisions are considered intrinsically asymmetric when cell fate determinants become asymmetrically segregated to the daughter cells during the division process. The mitotic spindle determines the plane of cell cleavage and plays a key role in the decision between symmetric and asymmetric cell division. Positioning of the mitotic spindle along the plane of a polarized epithelium supports symmetric division and addition of both daughter cells to the epithelial cell layer. Spindle positioning along the apicobasal axis of epithelial cells leads to asymmetric division and disconnects one of the daughter cells from the basal lamina. Hence the regulation of spindle positioning is critical for the orientation and plane of cell cleavage, as well as the choice between symmetric and asymmetric cell division.
The early Caenorhabditis elegans embryo has emerged as a powerful model for studies of spindle positioning and asymmetric cell division (Galli and van den Heuvel, 2008; Gonczy 2008). Fertilization of the C. elegans oocyte triggers establishment of anterior–posterior polarity, which in turn directs asymmetric localization of cell fate determinants. During asymmetric division of the zygote, the mitotic spindle elongates in anaphase while it becomes asymmetrically displaced to the posterior. This movement coincides with oscillations (rocking) predominantly of the posterior aster, perpendicular to the antero-posterior axis. These coordinated processes result in the creation of a larger anterior blastomere (AB) and a smaller posterior blastomere (P1). Severing of the spindle midzone with a UV laser revealed that spindle positioning is determined by forces that pull from the cell periphery on astral microtubules (Grill et al., 2001). Placement of the spindle off-center results from asymmetry in these cortical pulling forces and depends on the polarity of the one-cell egg (Grill et al., 2001).
The dynein minus end–directed microtubule motor, in combination with disassembly of microtubule plus ends, generates cortical pulling forces (Couwenbergs et al., 2007; Kozlowski et al., 2007; Nguyen-Ngoc et al., 2007; Laan et al., 2012). Dynein is recruited to the cell periphery by a conserved protein complex that consists of the LIN-5 coiled-coil protein (NuMA in mammals), the TPR-GoLoco motif proteins GPR-1 and GPR-2 (hereafter referred to as GPR-1/2, related to Drosophila Pins and mammalian AGS3 and LGN), and the α subunit of a heterotrimeric G protein (GOA-1 and GPA-16 in C. elegans, hereafter referred to as Gα; Colombo et al., 2003; Gotta et al., 2003; Srinivasan et al., 2003). GDP-bound Gα associates with the C-terminal GoLoco/GPR motifs of GPR-1/2, while the N-terminal TPR motifs of GPR-1/2 bind LIN-5 (Srinivasan et al., 2003; Afshar et al., 2004; Du and Macara, 2004; Fisk Green et al., 2004). Thus Gα proteins contribute to spindle positioning in a noncanonical, presumably receptor-independent manner. While lipid modification of the Gα proteins likely contributes to plasma membrane localization, it is currently unknown whether additional proteins associated with the membrane or the actin-rich cortex underneath promote Gα–GPR–LIN-5–dynein recruitment.
We report here our study of the membrane localization of Gα–GPR–LIN-5 and possible contribution of the cell cortex and transmembrane proteins in the generation of pulling forces. We show that localization of the LIN-5 complex at the cell periphery does not require cortical actin. Instead, severe reduction of cortical actin increased the net pulling forces that act on the anterior spindle aster. Thus anterior enrichment of actin during polarity establishment appears to contribute to polarized spindle pulling, possibly by increasing cortical rigidity (Kozlowski et al., 2007; Afshar et al., 2010; Redemann et al., 2010). We identified the transmembrane and coiled-coil domain protein TCC-1 through coimmunopurification with GOA-1 Gα. TCC-1 localizes to the endoplasmic reticulum (ER) membrane, affects GPA-16 Gα distribution, and inhibits cortical pulling forces that act on astral microtubules. In yeast two-hybrid (Y2H) assays, we did not detect association between TCC-1 and the Gα–GPR–LIN-5 module; rather, TCC-1 interacted with UNC-116 kinesin-1. The TCC-1/UNC-116 interaction is physiologically relevant, as inhibition of unc-116 and tcc-1 each caused a specific and highly similar disruption of spindle translocation to the cortex in meiosis. Our data emphasize that proper spindle positioning not only requires plasma membrane–localized Gα–GPR–LIN-5–dynein, but also depends on additional factors that include actin distribution and ER-associated proteins.
RESULTS
LIN-5–GPR-1/2 localization at the cell periphery does not require microtubules or microfilaments
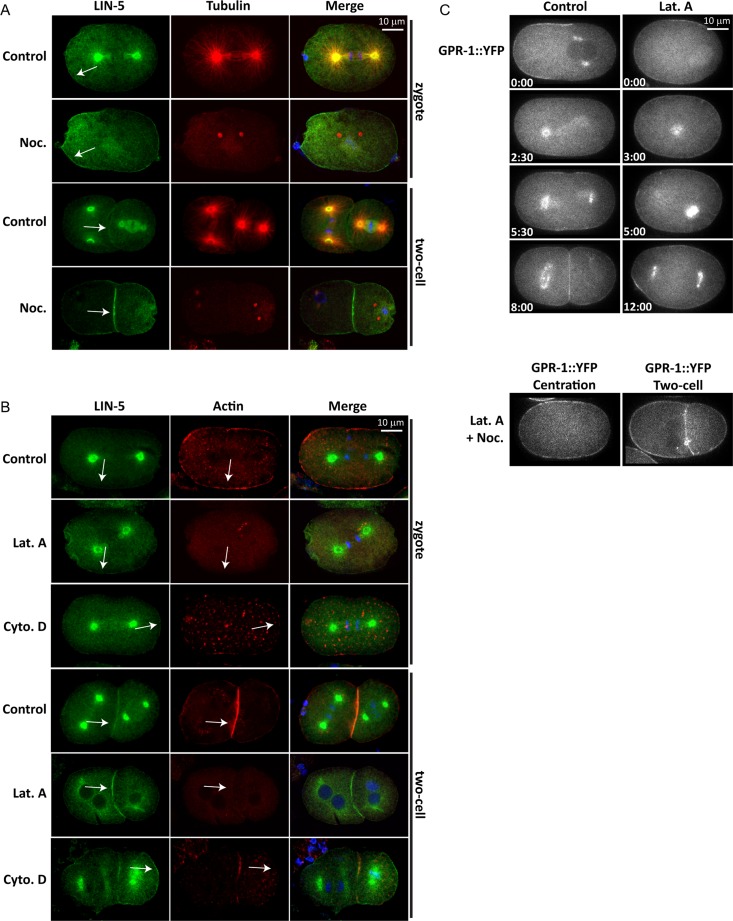
LIN-5 and GPR-1/2 colocalize at the spindle asters and cell periphery (Lorson et al., 2000). We have previously shown that localization of LIN-5 at the cell periphery is crucial for the generation of forces that pull astral microtubules to the cell cortex (van der Voet et al., 2009). We examined whether the peripheral localization of LIN-5 requires interaction with cortical actin or microtubules underneath the plasma membrane. For this purpose, we treated C. elegans embryos with drugs that interfere with the polymerization of microtubules (nocodazole) or actin (latrunculin A, cytochalasin D). C. elegans embryos are normally surrounded by an impermeable eggshell that prevents drug uptake. We used embryos from adults exposed to perm-1–feeding RNA interference (RNAi), as perm-1 RNAi causes the eggshell to be permeable, with no apparent effect on embryonic development (Carvalho et al., 2011). Brief treatment of perm-1(RNAi) embryos with nocodazole removed nearly all microtubules, as detected by immunohistochemistry (Figure 1A). This resulted in loss of LIN-5 from the spindle poles, but not from the cell periphery (Figure 1A). These observations are in agreement with previous studies (Lorson et al., 2000; Werts et al., 2011) and confirm that microtubules are not required for the localization of LIN-5 to the cortex or membrane. Treatment with latrunculin A drastically diminished the presence of cortical actin (Figure 1B), while actin was reduced to small spots at the membrane after cytochalasin D exposure. Nevertheless, LIN-5 remained associated with the cell membrane (Figure 1B). GPR-1 also remained membrane associated in the presence of latrunculin A, even after simultaneous disruption of microtubules and microfilaments (Figure 1C). Thus the cortical actin cytoskeleton does not appear necessary for localizing the LIN-5 complex to the cell periphery.
FIGURE 1:
Localization of the LIN-5 complex at the cell periphery does not require an intact actin or microtubule cytoskeleton. (A) perm-1(RNAi) embryos treated with the microtubule-depolymerizing drug nocodazole or with solvent only (1% DMSO, Control). Embryos were stained for LIN-5, tubulin, and DNA (DAPI). Note that microtubules are severely depolymerized in nocodazole-treated embryos, but that LIN-5 still localizes to the cell periphery (arrows). (B) perm-1(RNAi) embryos treated with the actin-depolymerizing drug latrunculin A, cytochalasin D, or with 1% ethanol (control for cytochalasin D). Embryos were probed with antibodies for LIN-5 and actin. DAPI was used to visualize DNA. Note that LIN-5 is present at the cell periphery of both control and latrunculin A–treated embryos (arrows). (C) Time-lapse images of YFP::GPR-1 embryos permeabilized with perm-1 RNAi and treated with solvent only (Control), latrunculin A, or a combination of latrunculin A and nocodazole. Note that even simultaneous disruption of microtubules and actin does not prevent GPR-1 membrane localization (bottom two sets of panels).
Actin inhibits pulling force generation in the anterior
Even though the actin cytoskeleton is not needed for localization of the LIN-5 complex, we noticed that embryos lacking intact actin filaments showed abnormal spindle behavior. During the first cell division in normal embryos, the mitotic spindle initially forms in the center of the cell. Asymmetric pulling forces cause spindle displacement toward the posterior during late metaphase and anaphase. Posterior movement of the spindle is accompanied by excessive rocking and flattening of the posterior spindle aster (Figure 2A and Supplemental Figure S1A). These spindle movements depend on the LIN-5–GPR–Gα complex, as inactivation of lin-5 and double inactivation of gpr-1 and gpr-2 or goa-1 and gpa-16 all reduce spindle elongation and eliminate spindle rocking, aster flattening, and cortical pulling forces (Srinivasan et al., 2003; Afshar et al., 2004; Nguyen-Ngoc et al., 2007; van der Voet et al., 2009).
FIGURE 2:
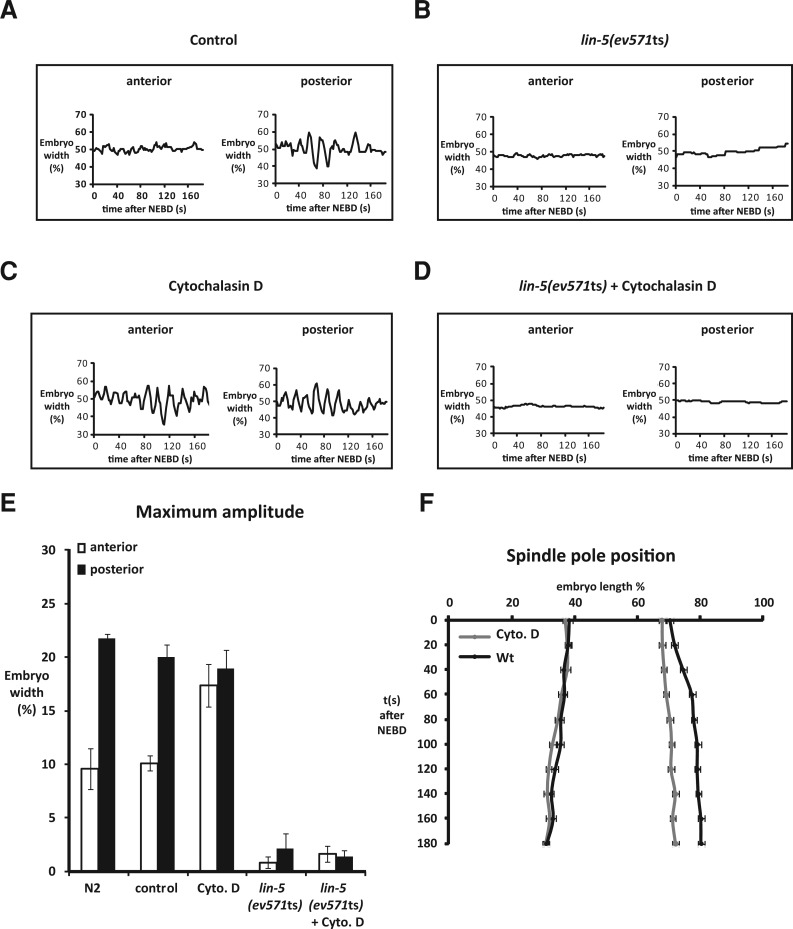
Actin inhibits pulling forces in the anterior. Centrosome movements are depicted for a control embryo (A), lin-5(ev571ts) embryo (B), cytochalasin D–treated embryo (C), and a lin-5(ev571ts) embryo treated with cytochalasin D (D). (E) Maximum amplitudes from spindle pole oscillations during anaphase. Values of the anterior and posterior spindle poles of indicated embryos are shown. Average values (± SD, n ≥ 6). (F) Position of the spindle poles during mitosis from nuclear envelope breakdown on in control and cytochalasin D–treated embryos (expressed as % egg length (x-axis) over time in seconds (y-axis), ± SD; control: n = 8; cytochalasin D: n = 9.
Flattening of the posterior spindle pole did not occur in embryos that lacked actin filaments after a brief cytochalasin D or latrunculin A treatment (Figure S1A). Treatment with these drugs also caused reduced elongation and lack of posterior displacement of the spindle in mitosis (Figures 2F and S1B). At the same time, disruption of the actin cytoskeleton enhanced rocking of the anterior spindle pole, as compared with untreated embryos (Figure 2, A, C, and E). Rocking of both the anterior and posterior spindle pole was abolished when we inactivated lin-5, by temperature shift of lin-5(ev571ts) embryos, in combination with cytochalasin D or latrunculin A treatment (Figure 2, B, D, and E). Thus, like the normal movements of the posterior pole, enhanced rocking of the anterior pole in the absence of filamentous actin depends on pulling forces generated by the LIN-5 complex. These data indicate that actin microfilaments contribute to the normal distribution of forces that pull on astral microtubule ends.
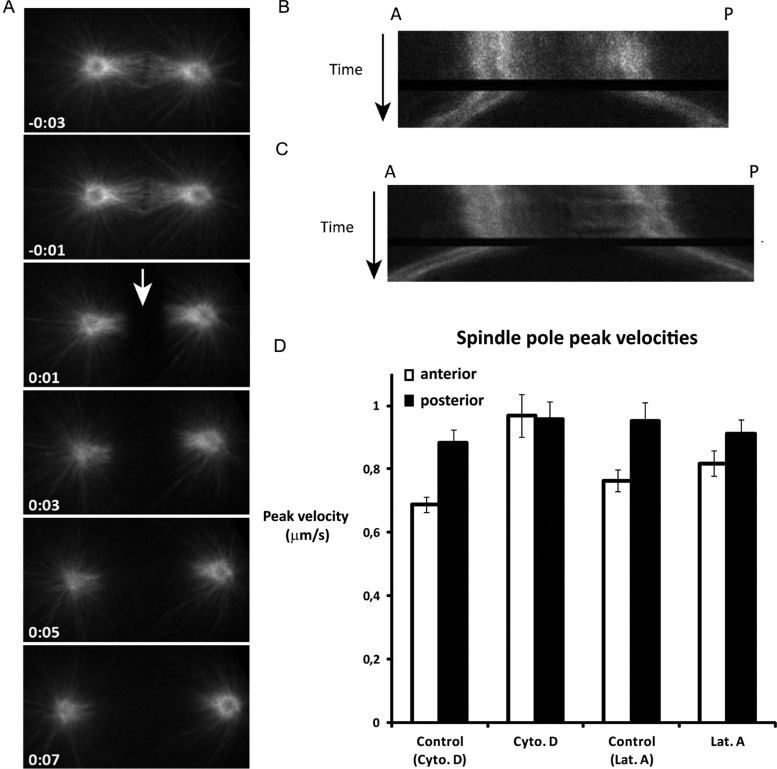
As only the anterior cortex contains a dense actin meshwork in early mitosis (Strome, 1986; Munro et al., 2004), disruption of the actin cytoskeleton would be expected to affect mostly anterior spindle pole behavior. The increased rocking of the anterior spindle pole appears to indicate that cortical actin normally inhibits the generation of pulling forces on astral microtubules. However, the reduced spindle elongation and lack of posterior centrosome flattening after actin disruption may point to a positive contribution of actin in spindle pulling forces. To assess whether actin stimulates or inhibits cortical pulling forces, we severed the midzone spindle at anaphase onset with a UV laser (Figure 3). Cytochalasin D treatment resulted in substantially increased pulling forces in the anterior, as deduced from the velocity of spindle pole movements after midzone severing (Figure 3, C and D, and Supplemental Movies S1 and S2). The pulling forces were similarly high in the anterior and posterior after treatment (Figure 3D), indicating that dense cortical actin normally antagonizes the net pulling forces in the anterior of the embryo. Latrunculin A treatment showed a similar trend (Figure 3D), although the velocity of the anterior pole remained just below the velocity of the posterior pole (Figure 3D).
FIGURE 3:
Actin inhibits anterior pulling force generation. (A) Time-lapse series of a mitotic spindle in a control embryo, in which the midzone spindle is severed at anaphase onset (arrow). Note that the posterior spindle pole moves with a higher velocity than the anterior spindle pole. (B and C) Kymographs of mitotic spindles from a control (B) and a cytochalasin D–treated embryo (C) after midzone severing. Kymographs are taken from a single longitudinal line across the mitotic spindle. The spindle poles are visualized by GFP::TBB-2. Compared with control embryos (B), both spindle poles move with high velocities after midzone ablation in cytochalasin D–treated embryos (C). (D) Spindle pole peak velocities after severing the spindle midzone at anaphase onset with a UV laser. Values are shown for embryos treated with cytochalasin D, latrunculin A, or solvent only (control Cyto. D: egg buffer + 1% EtOH; control Lat. A : egg buffer plus 1% DMSO). Pole velocities in the latter control embryos were slightly higher than normal (average values ± SEM). Anterior pole velocity is significantly increased in the presence of cytochalasin D (p < 0.0007 for solvent compared with cytochalasin D; p < 0.3 for solvent compared with latrunculin A. Pole velocities in the latter control embryos were slightly higher than normal. n ≥ 10 embryos for each treatment.
We considered whether loss of anterior–posterior polarity might explain the reduced asymmetry in pulling forces in these experiments. We started actin drug treatment after polarity establishment, but high pulling forces in the anterior are to be expected if the treatment somehow induces posteriorization. We followed PAR-2::green fluorescent protein (GFP) localization and observed that cortical polarity was maintained during the time course of our experiments (Figure S2), in agreement with results from others (Goehring et al., 2011). In conclusion, cortical actin is not needed for LIN-5 complex localization and pulling force generation at the cell periphery. In contrast, enrichment of the actin filament network at the anterior cortex during polarity establishment appears to reduce cortical pulling and contributes to the asymmetry in pulling forces that act on microtubule plus ends.
GOA-1 Gα appears the most critical membrane anchor of the pulling force complex
The results above indicate that the Gα–GPR-1/2–LIN-5 complex is directly linked to the plasma membrane, which agrees with results from other studies and the current paradigm in the field (Galli and van den Heuvel, 2008; Gonczy, 2008; Afshar et al., 2010; Redemann et al., 2010). One of these studies showed that weakening of the actin cytoskeleton during anaphase causes extensive invaginations of the cell membrane toward the spindle poles (Redemann et al., 2010). These invaginations probably reflect the localization of cortical force-generator complexes and illustrate substantial membrane attachment of the complex (Redemann et al., 2010). Lipid modification of the Gα subunit is probably critical for its membrane association, but it is unclear whether this provides the only membrane anchor for the complex.
We examined the contribution of Gα proteins GOA-1 and GPA-16 in LIN-5–GPR-1/2 localization and pulling force generation. While these Gα proteins act largely redundantly in spindle positioning (Srinivasan et al., 2003; Gotta et al., 2003; Colombo et al., 2003), they behave remarkably differently in several aspects (see Discussion; Afshar et al., 2004, 2005). When comparing wild-type embryos with the presumed null mutants goa-1(sa734) and gpa-16(ok2349)(Caenorhabditis Genetics Center [www.cbs.umn.edu/CGC]; Robatzek and Thomas, 2000), we observed a more substantial contribution of goa-1 to spindle pole oscillation, in particular in the absence of an intact actin cytoskeleton (Figure S3A), and to mitotic spindle elongation (Figure S4A). In addition, we observed that membrane invaginations in embryos with a disrupted actin cytoskeleton predominantly depend on goa-1. In control and gpa-16(ok2349) embryos treated with cytochalasin D, large invaginations were present during anaphase (Figure S3B). However, the number of cortical invaginations observed in cytochalasin D–treated goa-1(sa734) mutant embryos was remarkably decreased (Figure S3B). These results indicate that at a minimum GOA-1 Gα contributes to direct membrane anchoring of the pulling force complex.
TCC-1 affects spindle movements in the early C. elegans embryo
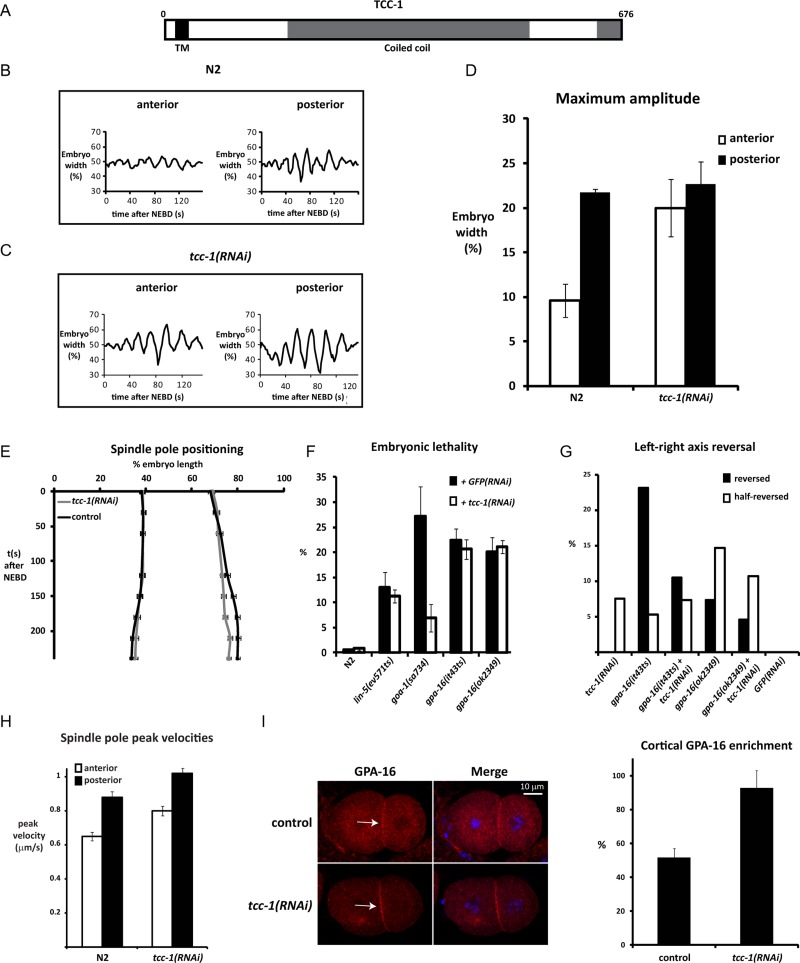
We hypothesized that additional membrane-associated proteins might contribute to the membrane anchoring of the Gα–GPR-1/2–LIN-5 complex. To identify such proteins, we immunopurified GOA-1 from embryonic lysates and characterized potentially associated proteins by mass spectrometry. GOA-1 purification enriched for known GOA-1 interaction partners, such as GPB-1 Gβ (16 peptides in bait, 0 in control), GPR-1 (3 peptides in bait, 0 in control), and RIC-8 (4 peptides in bait, 0 in control). Only a few proteins with putative transmembrane domains (as determined by TMHMM analysis; Transmembrane prediction program, based on a hidden Markov Model) were specifically copurified with GOA-1 (Table 1). To test whether these proteins contribute to spindle positioning, we performed RNAi of the corresponding genes in combination with time-lapse differential interference contrast microscopy of early embryos. While inhibition of catp-3 and eat-6 each caused early embryonic defects (unpublished data), Y59A8A.3 appeared to contribute more specifically to spindle behavior. The predicted Y59A8A.3 protein contains an N-terminal signal anchor motif followed by a single-pass transmembrane domain and extended coiled-coil region (Figure 4A). We named this protein TCC-1, for transmembrane and coiled-coil protein. tcc-1(RNAi) embryos displayed normal asymmetry in positioning of the cleavage plane (wild type, N2: 55.92 ± 2.19, n = 11; tcc-1(RNAi): 55.60 ± 1.81, n = 20; Figure S5). However, inhibition of tcc-1 by RNAi resulted in strongly increased spindle rocking (Figure 4, B–D) and reduced spindle elongation (Figure S4B). These spindle abnormalities suggested a contribution of TCC-1 in the generation or distribution of pulling forces.
TABLE 1:
Putative transmembrane proteins identified by mass spectrometry analysis of immunopurified GOA-1.
| Protein | Description | GOA-1 immunoprecipitation | Control immunoprecipitation |
|---|---|---|---|
| ANT-1.1 | Mitochondrial adenine nucleotide transporter | 8 | 0 |
| TCC-1 | Transmembrane and coiled-coil protein | 4 | 1 |
| ILE-1 | ERGIC-53 homologue | 2 | 0 |
| F01G4.6 | Putative phosphate carrier protein, mitochondrial | 2 | 0 |
| OSTB-1 | Oligo saccharyl transferase, β subunit | 1 | 0 |
| SRW-87 | Serpentine receptor, class W | 1 | 0 |
| SGCB-1 | Sarcoglycan, β dystrophin–associated glycoprotein | 1 | 0 |
| RME-2 | Low-density lipoprotein receptor | 1 | 0 |
| IMMT-2 | Inner membrane of mitochondria protein | 1 | 0 |
| F52G3.4 | 1 | 0 | |
| EAT-6 | α subunit of Na+/K+ ATPase | 1 | 0 |
| CATP-3 | Cation-transporting ATPase | 1 | 0 |
| C15H9.4 | 1 | 0 |
Numbers of peptides identified in the GOA-1 and negative-control immunoprecipitations are shown on the right.
FIGURE 4:
TCC-1 inhibits cortical pulling forces, possibly through inhibition of GPA-16 localization at the plasma membrane. (A) Illustration of TCC-1 domain structure. (B and C) Transverse spindle pole movements during anaphase in (B) a normal (N2) and (C) tcc-1(RNAi) embryo. (D) Maximum amplitude of the anterior and posterior spindle pole during anaphase in N2 and tcc-1(RNAi) embryos. Average values (± SD; n ≥ 10). (E) Spindle pole positioning in N2 and tcc-1(RNAi) embryos, indicated by the position of the centrosomes, expressed as % egg length (x-axis) over time in seconds (y-axis). (F) Embryonic lethality observed in the depicted strains treated with RNAi for gfp or tcc-1 at 20°C. Average values (± SD) from experiments performed in duplicate with three hermaphrodites for each condition. (G) Left–right axis reversal of indicated strains at 20°C, treated with RNAi for either gfp or tcc-1. Average values (± SD) from 65 to 112 animals were scored for each condition. (H) Spindle pole peak velocities after severing the midzone spindle at anaphase onset with a UV laser. Values are shown for normal (N2) and tcc-1(RNAi) one-cell embryos. Average values are indicated for each pole (± SEM; N2: tcc-1: n = 17; RNAi: n = 31). (I) Control and tcc-1(RNAi) embryos stained for GPA-16 and DNA (DAPI). Note that GPA-16 localization is enriched at the plasma membrane in tcc-1(RNAi) embryos (arrows). The right panel shows a quantification of cortical GPA-16 enrichment measured at the contact between the AB and P2 cell. Average values are indicated (± SD; n = 14).
Homology searches in other eukaryotes revealed significant protein similarity between TCC-1/Y59A8A.3 and various proteins in other species. However, identity in amino acid sequence is limited and largely restricted to the coiled-coil domain; even related nematode species show quite divergent TCC-1 sequences. This is a common aspect of coiled-coil proteins and renders homology predictions without substantial structural and functional analysis of little informative value.
TCC-1 lowers cortical pulling forces and plasma membrane association of GPA-16
To assess the relationship between TCC-1 and Gα in spindle movements, we examined the tcc-1 phenotype and studied its genetic interaction with goa-1 and gpa-16. Comparing tcc-1 and goa-1 loss-of-function embryos did not reveal a simple positive or antagonistic interaction between TCC-1 and GOA-1. Similar contributions of tcc-1 and goa-1 were only observed for anaphase spindle elongation. Similar to goa-1 knockdown, tcc-1 RNAi reduced mitotic spindle elongation in wild-type and gpa-16(ok2349) embryos but not in goa-1(sa734) embryos (Figures 4E and S4, A–D). Unlike goa-1 mutant embryos, however, rocking of the spindle was substantially higher than normal in tcc-1(RNAi) embryos, which was most apparent for the anterior pole (Figure 4, B–D). We performed spindle-severing experiments to obtain a more direct measure of pulling forces in tcc-1(RNAi) embryos. After the spindle midzone was cut with a UV laser at anaphase onset, both the anterior and posterior spindle poles in tcc-1(RNAi) embryos moved with significantly higher velocities toward the cortex than did the spindle poles in normal control embryos (Figure 4H; anterior: p < 0.002; posterior: p < 0.004). Thus, directly or indirectly, TCC-1 inhibits the net pulling forces between membrane-localized GOA-1/GPA-16 Gα proteins and plus ends of astral microtubules.
The observed effect in pulling forces might indicate that tcc-1 normally antagonizes goa-1 and/or gpa-16 Gα function. RNAi of tcc-1 suppressed the embryonic lethality associated with a goa-1 candidate null mutation, which indicates that tcc-1 acts at least partly independently of goa-1 (Figure 4F). Interestingly, we found that tcc-1 affects a highly specific function of gpa-16 in the maintenance of left–right asymmetry of the body (Bergmann et al., 2003). In normal four-cell embryos, the spindles of the ABa and ABp blastomeres undergo a highly reproducible skewing, which generates a characteristic left–right asymmetry during embryonic and adult development (Bergmann et al., 2003). As prominent features of this asymmetry, the anterior gonad arm is situated on the right side of the animal, while the posterior gonad arm is positioned at the left side. Conversely, the intestine becomes asymmetrically placed to the left in the anterior and to the right side in the posterior. In agreement with previous results, we observed that a substantial fraction of gpa-16(it143ts) and gpa-16(ok2349) mutants were left–right reversed, while goa-1(sa734) mutants showed normal left–right asymmetry (Figure 4G). RNAi of tcc-1 substantially suppressed the left–right reversal of gpa-16(it143ts) partial loss-of-function mutants (Figure 4G). The null allele gpa-16(ok2349) showed only limited suppression. As a possible explanation, these results could indicate that TCC-1 acts as an inhibitor of GPA-16.
Increased GPA-16 function after tcc-1 RNAi could explain rescue of embryonic lethality of goa-1(sa734) mutants and of restoration of left–right asymmetry in gpa-16(it143ts) mutants. We used immunostaining of early embryos to examine whether tcc-1 knockdown alters the expression or localization of GPA-16, GOA-1, or LIN-5. Downregulation of tcc-1 did not lead to a significant change in GOA-1 or LIN-5 localization (Figure S6). However, tcc-1(RNAi) embryos showed altered distribution of GPA-16, with increased localization to the cell periphery and/or reduced presence in the cytoplasm (Figure 4I). Together these data support the model that TCC-1 inhibits membrane-associated microtubule pulling forces at least in part by restricting cell membrane localization of GPA-16 Gα.
TCC-1 localizes to ER membranes
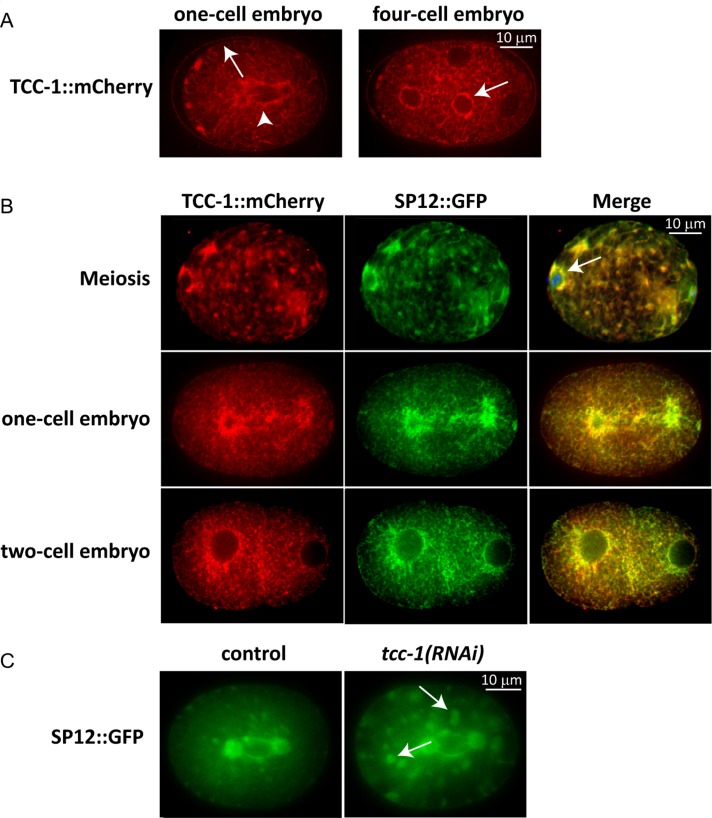
Because TCC-1 has a predicted transmembrane domain, the protein might be localized to the outer plasma membrane or to internal membranes. To assess the subcellular localization of TCC-1, we constructed a transgene for expression of a TCC-1::mCherry protein fusion under the control of the tcc-1 promoter and 3′-UTR sequences. We used the MosSCI technique to integrate a single-copy TCC-1::mCherry transgene on chromosome I (Frokjaer-Jensen et al., 2008). TCC-1::mCherry expression was observed throughout development, including the earliest stages of embryogenesis, in agreement with the tcc-1 RNAi phenotype (Figure 5, A and B; unpublished data). TCC-1::mCherry was present at cytoplasmic membranes that organized into a network with strong resemblance to the ER (Poteryaev et al., 2005). Similar as described for ER markers, TCC-1::mCherry localization surrounded nuclei, was visible along tubular membranes that extended throughout the cytoplasm, and associated with the plasma membrane (Figure 5, A and B). This network appeared to be highly dynamic and reorganized during mitosis, again as described in the ER (Poteryaev et al., 2005). To further examine whether TCC-1 localizes to the ER, we used the ER-resident signal peptidase SP12 fused to GFP as a marker for the ER (Poteryaev et al., 2005). Expression of TCC-1::mCherry and SP12::GFP within the same embryos confirmed the strong overlap between TCC-1::mCherry localization and the ER (Figure 5B). We wondered whether TCC-1::mCherry might get trapped along a normal trafficking route of TCC-1 to the plasma membrane. This possibility appears unlikely, as we found that TCC-1 contributes to the distribution of the ER. Compared with normal embryos (Movie S3), tcc-1(RNAi) embryos showed increased numbers and enlarged foci or aggregates in mitosis and also seemingly aggregated bundles that extend to the cortex (Figure 5C and Movie S4). In tcc-1(RNAi) embryos 12.2 ± 5.5 aggregates were observed at nuclear envelope breakdown (NEBD), compared with 3.2 ± 2.9 in control embryos. Extensive aggregation of ER structures was also seen after interference with homotypic membrane fusion or RAB-11 function in previous studies (Poteryaev et al., 2005, Zhang et al., 2008). Thus TCC-1 appears necessary for normal ER distribution, which fits well with the observed ER localization of TCC-1::mCherry. We conclude that TCC-1 localizes to the ER membrane and contributes to the dynamic organization of the ER in mitosis.
FIGURE 5:
TCC-1 localizes to the ER. (A) One-cell embryo at anaphase onset, and four-cell embryo in interphase. Note that TCC-1::mCherry associates with the mitotic spindle (left panel, arrowhead), nuclear envelope (right panel, arrow), and plasma membrane (left panel, arrow). The organization is more sheet-like in mitosis and becomes dispersed at the end of mitosis. (B) Localization of TCC-1::mCherry and the ER marker SP12::GFP in early embryos. Note the overlap in localization and close association of TCC-1::mCherry and SP12::GFP with the meiotic spindle and nearby cortex (top, arrow). The organization changes from membranous with foci in meiosis and mitosis to dispersed in interphase. (C) Extensive clustering of the ER in tcc-1(RNAi) embryo (right, indicated by arrows). In a wild-type embryo, some clusters can be detected in mitosis (left).
TCC-1 functions with UNC-116 kinesin-1
As a first step in identifying the molecular function of TCC-1, we performed a Y2H screen in search for TCC-1–interacting proteins. For this purpose, we generated eight different TCC-1 baits that included the full-length protein and various overlapping fragments (see Materials and Methods for details). This strategy resulted in identification of a series of candidate TCC-1–interacting proteins, as summarized in Table 2. Among the most frequent hits were the kinesin-1 heavy chain UNC-116, the dynactin subunit DNC-2 Dynamitin, and the adaptor protein Spindly, which recruits dynein to kinetochores (Gassmann et al., 2010). Both kinesin-1 and dynein/dynactin motor complexes contribute to spindle positioning in the early embryo (Yang et al., 2005; Couwenbergs et al., 2007; Nguyen-Ngoc et al., 2007; van der Voet et al., 2009). DNC-2 is found relatively frequently in Y2H screens, therefore any interactions involving DNC-2 should be interpreted with caution. Nevertheless, the interactions of TCC-1 with three motor complex–related proteins suggest a direct link between TCC-1 and microtubule-based motor proteins.
TABLE 2:
Potential TCC-1–interacting proteins identified in a Y2H screen.
| Gene name | Description | Hits |
|---|---|---|
| daf-21 | Hsp90 family member | 51 |
| spdl-1 | Spindly | 34 |
| unc-116 | Kinesin heavy chain 1 | 8 |
| dnc-2 | Dynactin complex component (dynamitin) | 8 |
| F35C11.5 | 5 | |
| ntl-3 | NOT-like (yeast CCR4/NOT complex component) | 4 |
| eftu-2 | Elongation factor TU family | 4 |
| dlg-1 | Discs large | 3 |
| Y59A8A.3 | Y59A8A.3 | 3 |
| ubxn-2 | (ubiquitin regulatory X) domain-containing protein | 2 |
| flap-1 | Friend leukemia virus integration 1-Associated Protein homologue | 2 |
| Y59A8B.10 | 2 | |
| Y39B6A.1 | Related to human Hornerin | 2 |
| npp-9 | Nuclear pore complex protein | 2 |
| glp-1 | Notch receptor | 2 |
Note that daf-21 was only identified with baits that contained the TCC-1 transmembrane region, probably illustrating interaction with the exposed hydrophobic domain. dnc-2 is frequently found in Y2H screens and could be a promiscuous interaction.
Meiotic spindle positioning and rotation requires sequential kinesin-1 and dynein activity. We examined these processes as attractive functional readouts for kinesin-1 and dynein motor activity (Yang et al., 2005; Ellefson and McNally, 2009; van der Voet et al., 2009). Once the meiotic spindle is formed, the spindle and chromosomes undergo kinesin-1–dependent translocation to the cortex (Yang et al., 2005). Subsequently, upon activation of the APC/C complex, dynein activity rotates the meiotic spindle and orients the spindle perpendicular to the cortex (Ellefson and McNally, 2009; van der Voet et al., 2009; McNally et al., 2010). These combined movements are repeated in meiosis II, allowing expulsion of chromosomes from the cytoplasm into small polar bodies. As we identified kinesin-1 and DNC-2 as potential TCC-1–interacting proteins, we examined meiotic spindle positioning in tcc-1(RNAi) fertilized eggs.
We first examined TCC-1::mCherry localization in meiosis to visualize whether TCC-1 is present during meiotic spindle translocation and rotation. As seen in mitosis, TCC-1::mCherry and SP12::GFP were closely associated with the spindle at the stage of meiotic spindle translocation (Figure 5B). In addition, we observed clustering of the TCC-1::mCherry network throughout the cytoplasm and patches of ER in close proximity to the cell membrane (Figure 5B).
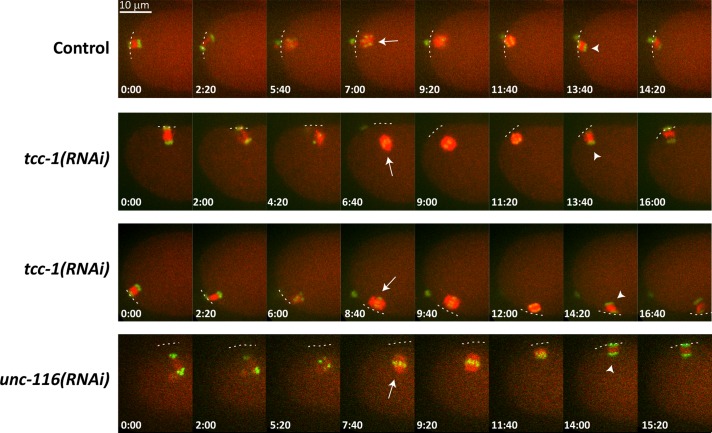
In control oocytes, the meiotic spindle moved to and remained in close proximity with the cortex throughout meiosis (12/12 oocytes; Figure 6, top row, and Movie S5). Furthermore, the meiotic spindle in all observed cases underwent dynein-dependent rotation. In contrast, tcc-1(RNAi) oocytes showed defects in meiotic spindle translocation (6/11 oocytes). In these cases, the meiotic spindle was either positioned at a significant distance from the cortex or spindles drifted toward the posterior during meiosis (Figure 6, second and third rows, and Movies S6 and S7). This behavior of the meiotic spindle after tcc-1 RNAi closely resembles the spindle movements in the fertilized egg following unc-116 RNAi (Figure 6, last row, and Movie S8). Even when meiotic spindles did not remain positioned in the anterior, the dynein-dependent rotation and membrane movement of the spindle during anaphase occurred as in the wild-type. Rotation even occurred in tcc-1, unc-116 double-RNAi embryos as opposed to lin-5(RNAi) embryos (Figure S7 and Movies S9–S11). Thus TCC-1 participates in the UNC-116–mediated translocation of the meiotic spindle, but not detectably in spindle rotation mediated by ASPM-1, LIN-5, and dynein (van der Voet et al., 2009). In summary, identification of UNC-116 kinesin-1 and TCC-1 as binding partners in Y2H screens and the similar loss-of-function phenotypes of unc-116 kinesin-1 and tcc-1 in meiosis indicate that TCC-1 and kinesin-1 interact in vivo to translocate the meiotic spindle toward the cortex.
FIGURE 6:
TCC-1 contributes to translocation of the meiotic spindle toward the cortex. Depicted are still images from time-lapse recordings of normal (Control), tcc-1(RNAi), and unc-116(RNAi) embryos. In control embryos, the meiotic spindle stays in close proximity to the anterior cortex. After spindle shortening, a rotation causes the spindle to be positioned perpendicular to the cortex (arrowhead). In tcc-1(RNAi) embryos, the meiotic spindle becomes transiently displaced from the cortex (arrow, second row), as in unc-116(RNAi) embryos (arrow, bottom row). In some tcc-1(RNAi) embryos, the meiotic spindle appears to drift or tumble away from the anterior end (arrow, third row). See Supplemental Movies S1–S11 for further details.
Kinesin 1 has been implicated in meiotic but not mitotic spindle positioning in C. elegans. Because of its functional link to tcc-1, we examined whether unc-116 RNAi also affects mitotic spindle movements. We observed a mild but significant increase in aster movements during mitotic anaphase of the zygote (Figure S8). We propose that the TCC-1/UNC-116 interaction helps to link the ER to spindle microtubules, which promotes translocation of the meiotic spindle, yet dampens oscillations of the mitotic spindle. Rocking was more substantially increased in tcc-1(RNAi) embryos, which probably corresponds to the increased fraction of GPA-16 Gα at the plasma membrane of such embryos.
DISCUSSION
In this study, we investigated whether factors in addition to Gα–GPR–LIN-5–dynein contribute to the pulling forces that position the mitotic spindle in the asymmetrically dividing C. elegans zygote. We initially considered proteins associated with the cortex, the actin-rich layer at the inner face of the plasma membrane. We found that actin filaments are not required for localization of the LIN-5 complex or pulling force generation and that actin in fact antagonizes the forces that pull on astral microtubules. These results are consistent with and may be explained by observations from other studies (see below, e.g., Kozlowski et al., 2007; Afshar et al., 2010; Redemann et al., 2010). While we attempted to block actin assembly completely, a previous study focused on weakening the cell cortex, by partial nmy-2 RNAi or interfering with microfilaments (Redemann et al., 2010). These conditions also revealed the maintenance and anterior increase in pulling forces, but also the inward pulling of long membrane invaginations toward the centrosome. This indicated that shrinking microtubules can remain attached to the membrane for prolonged times, which we found to depend critically on the Gα subunit GOA-1. In the second part of this study, we investigated whether, in addition to lipid anchoring of the Gα subunit, membrane-associated proteins can be identified that contribute an anchor function for the pulling force generators. We identified TCC-1, a predicted transmembrane protein, which contributes to multiple aspects of spindle positioning. While a functional interaction between TCC-1 and GOA-1 remains to be established, we found TCC-1 to affect spindle movements in meiosis and mitosis. A role of TCC-1 in the cytoplasmic retention of GPA-16 Gα and a putative role as a kinesin-1 adaptor at the ER membrane probably both contribute to the observed effects in spindle behavior.
During polarity establishment, cortical actomyosin retracts from the posterior and accumulates in the anterior of the embryo (Strome, 1986; Munro et al., 2004). A negative role for actin in pulling force generation was anticipated in modeling studies of spindle oscillation and posterior displacement (Kozlowski et al., 2007). In these studies, increased stiffness of the cortex (cortical rigidity) was considered likely to result in reduced force generation, as it increases the rate of detachment of the force generator from the shrinking microtubule. Increasing cortical rigidity in the simulations dampened spindle pole oscillations (Kozlowski et al., 2007). Structural components of the cortex, such as actin, likely contribute to cortical rigidity, and the extensive enrichment of actomyosin in the anterior cortex should lead to increased stiffness. Thus the observations that removal of filamentous actin leads to increased anterior spindle pole oscillations and increased pulling forces in the anterior fully agree with computer simulations by others (Kozlowski et al., 2007).
Disrupting actin polymerization obviously has effects other than creating a more flexible cortex. In particular, the contribution of actin in polarity maintenance should be considered. Actomyosin is critical for polarized PAR protein localization (Munro et al., 2004; Severson and Bowerman, 2003). In addition to actomyosin reorganization as a trigger for anterior–posterior polarity establishment, actin-dependent endocytosis is higher in the anterior of the zygote and helps maintain PAR-6 levels in the anterior (Nakayama et al., 2009; Shivas and Skop, 2012). Nevertheless, a previous study carefully examined permeabilized embryos treated with latrunculin A and cytochalasin D and demonstrated maintenance of PAR localization and dynamics during short-term microfilament disruption (Goehring et al., 2011). We followed a very similar procedure and did not observe polarity loss during the time course of our experiments. Hence we consider reduced cortical stiffness, and not PAR protein mislocalization, as a probable cause for increased rocking and velocity of the anterior pole.
The effects of physical properties such as cortical rigidity and cell shape in spindle positioning can be overruled by localized activity of pulling force generators. For instance, during centration-rotation, the maternal and paternal pronuclei migrate together toward the actomyosin-enriched anterior cortex. This movement corresponds to a greater number of LIN-5 pulling force complexes in the anterior in early mitosis (Park and Rose, 2008; Galli et al., 2011) and probably also involves dynein attached to intracellular organelles (Kimura and Kimura, 2011). Anterior-directed movement of the pronuclei stops at the time of nuclear envelope breakdown, through inhibitory phosphorylation of LIN-5 by the atypical protein kinase C PKC-3 (Galli et al., 2011). Subsequently, and even in the absence of this phosphorylation, the spindle becomes displaced to the posterior in anaphase. These observations confirm that multiple factors contribute spatiotemporal control of spindle positioning. This may include regulated localization of the pulling force generators, for instance, through asymmetric LET-99 localization, GPB-1 trafficking, and/or PPK-1 kinase activity (Tsou et al., 2002; Panbianco et al., 2008; Thyagarajan et al., 2011). Importantly, the observed effects of actin depletion on spindle pole velocity indicate that localized enrichment of actomyosin may be a major determinant in asymmetric positioning of the mitotic spindle, probably through its effects on cortical rigidity.
With some similarity to actin depletion, down-regulation of tcc-1 also increased spindle rocking and net pulling forces. However, in contrast to actin filaments, TCC-1 appeared to inhibit anterior- as well as posterior-directed forces. We identified TCC-1 as a protein coimmunopurified with GOA-1. TCC-1 and GOA-1 contributed similarly to spindle elongation in anaphase, yet tcc-1 RNAi suppressed the embryonic lethality associated with a goa-1–null mutation. This discrepancy might result from inhibition of GPA-16 by TCC-1. Despite their redundant contribution in spindle pulling, GOA-1 and GPA-16 behave remarkably differently in multiple functional assays. For instance, only GOA-1 interacts with GPR-1/2 in Y2H assays (Gotta et al., 2003; unpublished data), only GPA-16 has been found to contribute to left–right asymmetry (Bergmann et al., 2003), and GOA-1 and GPA-16 are differently regulated by RIC-8 (Afshar et al., 2004, 2005). RIC-8 acts as a nonreceptor guanine nucleotide exchange factor (GEF) for GOA-1 but not for GPA-16. In contrast, RIC-8 stimulates GPA-16 protein levels and plasma membrane localization, while it does not affect GOA-1 protein levels (Afshar et al., 2005). Similar observations are made for mammalian RIC-8 proteins, which, apart from their role as GEFs, appear to act as chaperones for specific subsets of Gα proteins (Gabay et al., 2011). As a possible mechanism, TCC-1 might counteract interaction of GOA-1 and GPA-16 with RIC-8. This could explain a positive effect on GOA-1, by promoting the GDP-bound state that interacts with GPR-1/2, while at the same time inhibiting plasma membrane localization of GPA-16. Alternatively, TCC-1 may simply affect the trafficking of GPA-16 but not GOA-1. Gα proteins localize to the plasma membrane through an intracellular trafficking route that involves the ER (Michaelson et al., 2002; Marrari et al., 2007). Thus localization of TCC-1 at the ER membrane is consistent with Gα protein interaction and effects in GPA-16 trafficking.
TCC-1 was previously identified in a Y2H screen for binding partners of the UNC-83 KASH domain protein (Fridolfsson et al., 2010). Similar to TCC-1, UNC-83 was found to interact with components of kinesin-1, as well as dynein motor complexes, and to act together with kinesin-1 and dynein in bidirectional nuclear migrations (Fridolfsson et al., 2010). Simultaneous binding to kinesin and dynein complexes promotes the transport of multiple organelles within the cell (Welte, 2004). The screen for UNC-83 interactors also identified Bicaudal D, a well-known adaptor protein that links dynein motors to vesicles but also interacts with antagonistic kinesin-1 motors (Splinter et al., 2010). Such observations support the existence of organelle-specific linker proteins that recruit antagonistic microtubule motor complexes to facilitate bidirectional transport. Similar to nuclear migration (Fridolfsson and Starr, 2010), movement of the ER along microtubules involves kinesin-1 and dynein (Wozniak et al., 2009). Loss of TCC-1 led to defects in meiosis that mimicked loss of kinesin-1. While suggested by Y2H results, dynein-dependent rotation of the meiotic spindle remained normal in tcc-1(RNAi) embryos. Thus functional support for a TCC-1/dynein connection is currently missing. Hence we propose that TCC-1 acts as an ER and plasma membrane adaptor for kinesin-1 in C. elegans.
Although the contribution of kinesin-1 in meiotic spindle translocation has been well documented, several questions remain. Most importantly, the localization of kinesin-1 during spindle translocation is unclear, and the organization and orientation of microtubules at the cortex is not known during meiosis. The meiotic spindle in C. elegans does not contain centrioles or astral microtubules, and cytoplasmic microtubules may not be organized unidirectionally. We observed localization of TCC-1 in a highly dynamic tubular network throughout the cytoplasm and enriched along the cortex. Moreover, TCC-1 localized in close association with the spindle in meiosis, as in mitosis. This mimicked the reported localization of the ER and overlapped with the ER marker SP12::GFP (Poteryaev et al., 2005). Previous immunostaining experiments did not reveal any specific subcellular localization of kinesin-1 at this stage (McNally et al., 2010). We propose that ER-localized TCC-1 in close association with the meiotic spindle and cell membrane provides a kinesin-1 anchor for microtubule-dependent spindle translocation. In addition, TCC-1/kinesin-1–dependent ER movements may contribute to meiotic spindle translocation.
Positioning of the mitotic spindle is widely considered to depend on interactions between astral microtubules and the cell cortex. During mitosis of the C. elegans zygote, pulling forces are mediated by the LIN-5 complex in association with the minus end–directed dynein motor at the cell cortex (Nguyen-Ngoc et al., 2007; Galli and van den Heuvel, 2008). However, during centration, dynein complexes localized at cellular organelles appear to be a major source of force generation for anterior-directed centrosome movements (Kimura and Kimura, 2011). Dynein and kinesin complexes are localized throughout the cell (Gonczy et al., 1999; McNally et al., 2010) and mediate a variety of spindle and organelle movements. This study implicates TCC-1 as a novel linker for kinesin-1 and possibly dynein complexes. We propose that the TCC-1/UNC-116 interaction helps to connect the ER to spindle microtubules, which promotes translocation of the meiotic spindle to the cortex, yet may dampen spindle oscillations and pulling forces in mitosis. Removal of TCC-1 in mitosis alters the distribution of the ER, possibly through reduced microtubule interaction, and allows more prominent cortical localization of GPA-16 Gα. This combination explains the vigorous rocking of the mitotic spindle in tcc-1(RNAi) one-cell embryos, as well as the high net forces that pull at the spindle poles from the cell periphery in such embryos. Our results emphasize the complex interplay between force generators at multiple cellular locations and physical properties of cellular structures, which together accomplish processes such as organelle movement and spindle positioning.
MATERIALS AND METHODS
C. elegans strains and culture
Strains were cultured on nematode growth medium plates, seeded with Escherichia coli OP50. Animals were maintained at 20°C, unless stated otherwise. Strains used were N2, SV124 lin-5(ev571ts) (maintained at 15°C), SA250 unc-119(ed3)III;tjls54[Ppie-1::gfp::tbb-2, Ppie-1::2xmCherry::tbg-1, unc-119(+)]; tjls57[Ppie-1::mCherry::H2B], RB1816 gpa-16(ok2349) (maintained at 15°C), BW1809 gpa-16(it143ts) (maintained at 15°C), DG1856 goa-1(sa734) (maintained at 15°C), RM1702 ric-8(md303), DS98 mat-2(ax102ts) (maintained at 15°C), KK866 itls153[pMW1.03 Ppie-1::gfp::par-2 pRF4], WH327 unc119(ed3);ojls23[pie-1::gfp::C34B2.10] (a gift from A. Spang, University of Basel), and SV1317 (heSi101[Ptcc-1::tcc-1::mCherry::tcc-1 +cb unc-119]I; unc-119(ed3) III). SV1317 was constructed by making use of the MosSCI technique (Frokjaer-Jensen et al., 2008).
Molecular cloning
We generated a TCC-1-mCherry construct (Ptcc-1::tcc-1::mCherry::tcc-1 3′ UTR) for single-copy chromosome I integration, using pCFJ352 (a gift from E. Jorgensen, Howard Hughes Medical Institute, University of Utah) as a vector backbone. The 2-kb region upstream of the tcc-1 start codon was used as a promoter sequence, and a 448–base pair fragment downstream of the stop codon was used as 3′ UTR. A fusion was made of 4106 base pairs of genomic tcc-1 sequences from the start codon to the ScaI site of exon 4, combined with a tcc-1 cDNA fragment starting at the ScaI site until, but not including, the stop codon. The mCherry coding sequence (a gift from R. Tsien, Department of Chemistry and Biochemistry, University of California, San Diego) was inserted in frame at the 3′ end of tcc-1. All fragments were obtained by PCR using KOD polymerase (EMD Millipore, Billerica, MA). Sequences are available upon request.
For Y2H assays, TCC-1 fragments were generated from cDNA using KOD polymerase and cloned into bait vector pMB28. Fragments were generated by using all possible combinations of the following primers: 5′-atggactcgtcattcgtcga-3′, 5′-aatcgacaagaggataattttg-3′, 5′-caattcttggctggagcatt-3′, 5′-ggattggtgatttgtgagag-3′, 5′-ttatgccaattgtgctctcag-3′, 5′-cggttcggcttctgtcttt-3′, and 5′-aatataagcttcatcgagttc-3′.
RNAi
For obtaining tcc-1(RNAi) and perm-1(RNAi) embryos, either young adults were injected with double-stranded RNA or L4-stage larvae were put on feeding plates with corresponding RNAi bacteria. Feeding plates were prepared as described elsewhere (Sato et al., 2009). RNAi clones from the Ahringer and Vidal libraries were used (Kamath et al., 2003; Rual et al., 2004). Embryos were dissected from adults ∼48 h (tcc-1) or 20 h (perm-1) after the onset of RNAi treatment.
Drug treatment of C. elegans embryos
For disruption of the actin or microtubule cytoskeleton in perm-1(RNAi) embryos, adults were splayed in 0.8× egg salt (containing 94 mM NaCl, 32 mM KCl, 2.7 mM CaCl2, 2.7 mM MgCl2, 4 mM HEPES, pH 7.5; Tagawa et al., 2001) in which latrunculin A (1 mM), cytochalasin D (20 μg/μl), nocodazole (10 μg/ml), ethanol (1%, control for cytochalasin D treatment), or dimethyl sulfoxide (DMSO; 0.5%, control for latrunculin A and nocodazole treatments) was added. For immunostaining of drug- or solvent-treated perm-1(RNAi) embryos, adults were splayed on glass slides in 50 μl of the indicated solutions, prepared as stated above. After dissection of the embryos, slides were incubated at room temperature in moist chambers for 5 min. Next the embryos were transferred in aliquots of 10 μl to poly-l-lysine–coated slides. After the embryos were transferred, coverslips were placed on the slides, and the embryos were freeze-cracked, fixed, and immunostained as described in the following section. For live imaging of treated perm-1(RNAi) embryos, a single adult worm was splayed in 2.5 µl of the indicated solutions on a coverslip. Next coverslips were attached to 1-mm-thick concave slides (Vermandel, Hulst, The Netherlands) to avoid pressure on the embryos. Coverslips were attached and sealed, by applying oil (Halocarbon, New York, NY) on the edges of the concave area. Only embryos that were at the stage of pronuclear meeting when dissected from adults were used for further analysis. They were imaged from NEBD onward (∼2 min later) for several minutes.
Antibodies and immunofluorescence staining
For immunostaining of embryos other than perm-1(RNAi), embryos were dissected from adults in 10 μl of water on poly-l-lysine–coated slides. Embryos were freeze-cracked and fixed for 5 min in methanol at −20°C and then for 20 min in acetone at −20°C. WH327 embryos were freeze-cracked and fixed for 15 min at −20°C in a solution containing 75% methanol, 4% formaldehyde, 0.5× PBS, and 50 mM EDTA; this was followed by fixation in methanol at −20°C for 5 min. After fixation, the embryos were rehydrated in phosphate-buffered saline (PBS) containing 0.05% Tween-20 (PBST) and blocked with blocking solution (PBST containing 1% bovine serum albumin and 1% goat serum [Sigma Aldrich, St. Louis, MO]) for 1 h. Embryos were stained with primary and secondary antibodies for 1 h and washed after each incubation with PBST four times, 15 min each time. Finally, the embryos were embedded in ProLong Gold Antifade containing 4′,6-diamidino-2-phenylindole (DAPI). Primary antibodies used in this study were: rabbit anti–LIN-5 (1:100), mouse anti–LIN-5 (1:10; Lorson et al., 2000), mouse anti-tubulin (1:400; Sigma-Aldrich), rabbit anti–GPA-16 (1:100; a gift from P. Gönczy; Afshar et al., 2005), mouse anti-actin (1:100) (MP Biomedicals, Santa Ana, CA), mouse anti-GFP (Roche Applied Science, Penzberg, Germany), and mouse anti-DsRed (Clontech Laboratories, Mountain View, CA). Secondary antibodies were used at a concentration of 1:500. Secondary antibodies used were: goat anti-rabbit Alexa Fluor 568, goat anti-mouse Alexa Fluor 568, goat anti-rabbit Alexa Fluor 488, and goat anti-mouse Alexa Fluor 488 (Invitrogen, Life Technologies, Carlsbad, CA). Images were taken with a 100×/1.4 numerical aperture (NA) lens or 60×/1.4 NA (Figure 5B, top row) on a Zeiss confocal microscope. Images depicted in Figure 5B (second and third rows) were taken with a 100×/1.4 NA lens on a Nikon spinning-disk microscope.
Time-lapse and live-cell imaging
For time-lapse imaging, embryos and oocytes were dissected in 0.8× egg buffer on coverslips, and slides were prepared as described above. Spindle oscillations and elongation were examined in embryos of the indicated strains at 20°C, unless stated otherwise. From NEBD onward, images were taken with Nomarski optics at 2.5-s intervals. Relative position of the spindle poles was measured in ImageJ. Embryos of strain SA250 were examined for spindle pole flattening during anaphase. From NEBD onward, images were taken by making use of Nomarski optics at 20-s intervals. GFP::PAR-2 distribution was examined in wild-type or perm-1(RNAi)–treated KK866 embryos in 0.8× egg salt. perm-1(RNAi)–treated KK866 embryos were either subjected to cytochalasin D (20 μg/μl) or 1% ethanol (control). Images were taken with a 100×/1.4 NA lens on a Nikon spinning-disk microscope at 30-s intervals. For envisioning cortical invaginations during metaphase and anaphase, perm-1(RNAi) embryos were dissected on coverslips and attached to concave slides as described above, using 0.8× egg buffer containing 1 μg/μl cytochalasin D and 0.5 μg/μl FM 1-43FX (Molecular Probes, Life Technologies, Carlsbad, CA) to visualize the plasma membrane. Images were taken at 2-s intervals for 3 min from NEBD onward. TCC-1::mCherry was visualized in SV1317 embryos by recording single images at the depicted stages of embryonic development. ER dynamics in control and tcc-1(RNAi) embryos were determined by making use of WH327 embryos. Images were taken at 30-s intervals with a 100×/1.4 NA lens Zeiss microscope. Analysis of meiotic spindle translocation and rotation was performed in SA250 embryos, either untreated or treated with RNAi for tcc-1, unc-116, lin-5, tcc-1+unc-116, or tcc-1+lin-5. After dissection from the adults, images were taken at 20-s intervals until completion of meiosis. All images were taken with a 100×/1.4 NA lens on a Nikon spinning-disk microscope. Images were processed by making use of ImageJ or Adobe Photoshop.
Quantification of cortical fluorescence
For quantifying cortical intensity of GOA-1, GPA-16, and LIN-5 staining, confocal images were taken of embryos at the two-cell stage. Cytoplasmic intensities were determined by averaging the intensities from a large cytoplasmic area of the anterior and posterior cell. The centrosomal region was avoided. For cortical fluorescence, intensity profiles of five 2 μm2 areas were measured that crossed the cortex between the AB and P2 cell at equally spaced distances. The average of the peak values was used. Fluorescence intensities were measured in ImageJ. Line scan analysis of GFP::PAR-2 distribution was performed by measuring fluorescence intensities in KK866 embryos in a line encompassing the anterior cortex, cytoplasm, and posterior cortex. The line width used was 2 μm.
Spindle severing
Ablation of the midzone was performed as described elsewhere (Grill et al., 2001). Laser ablations were carried out on a spinning-disk confocal microscope system, based on an inverted Nikon Eclipse Ti-E research microscope (Nikon) with a perfect focus system (Nikon), CFI S Fluor 100×/0.5–1.3 NA oil objective (Nikon), CSU-X1-A1 Spinning Disc (Yokogawa), and a Photometrics Evolve 512 EMCCD camera (Roper Scientific), controlled with MetaMorph 7.7 software (Molecular Devices). Sixteen-bit images were projected to the camera chip with an intermediate 2.0× lens (Edmund Optics) at a magnification of 0.066 μm/pixel. The microscope was equipped with a custom-ordered illuminator (MEY10021; Nikon) modified by Roper Scientific France/PICT-IBiSA, Institute Curie. A 491-nm (100-mW) Calypso (cobalt) laser was used for excitation. The spinning disk was equipped with a 405–491–561 triple-band mirror and an ET-GFP emission filter (Chroma). For the photoablation experiments, we used an ILas system (Roper Scientific France/PICT-IBiSA, Institut Curie) installed on the microscope. A 355-nm passively Q-switched pulsed laser (repetition rate: 6 kHz; energy/pulse: 2.5 mkJ; average power: 13 mW; pick power: 6 kW; pulse width: 400 ps; Teem Photonics) was used for the spindle severing. Images of GFP::TBB-2 embryos were taken at 0.5-s intervals at room temperature. Spindle poles were automatically tracked by making use of the MTrack2 plug-in in Fiji (Fiji Is Just ImageJ). Peak values of both the anterior and posterior pole were determined within a time frame of 10 s after ablation.
Characterization of embryonic lethality and left–right symmetry
L4 larvae of the indicated strains were placed on plates with RNAi feeding bacteria targeting either tcc-1 or GFP (control). Plates were held at 20°C. Animals were transferred to a new plate every day. For embryonic lethality scoring, plates were examined after 24 h. When progeny reached the young adult stage, left–right asymmetry was scored as described elsewhere (Bergmann et al., 2003).
Immunoprecipitations
For identification of GOA-1–interacting proteins, synchronized L1 DS98, mat-2 and DG1856 (control) worms were grown at 15°C in S medium containing HB101 bacteria. For obtaining embryos at an early stage of development, cultures were individually monitored, and embryo pellets were obtained from very young adults (DG1856). As described elsewhere (Galli et al., 2011), DS98 liquid cultures were shifted to 25°C for 2 h when most of the worms had just reached young adult stage. Thereafter, liquid cultures were cooled to 15°C, and the worms were allowed to grow for an additional 30 min. Embryos were obtained by hypochlorite treatment of the adult worms. Embryo pellets were ground two times for 30 s at a frequency of 1500/min in a Mikro-Dismembrator (Sartorius, Goettingen, Germany). Ground embryo pellets were lysed in lysis buffer (20 mM Tris-HCl, pH 7.8, 250 mM NaCl, 1% Triton X-100, 0.5 mM EDTA, 1 mM β-mercaptoethanol, 100 μM GDP, 10 mM 1-naphthyl phosphate monosodium salt monohydrate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate decahydrate, 100 μM sodium orthovanadate and protease inhibitors [complete, Mini, EDTA-free; Roche]) for 15 min at 4°C. The lysate was cleared at 13,000 rpm for 15 min at 4°C. Immunoprecipitations were performed with 15 mg of total protein with 30 μl rabbit anti–GOA-1 antibodies (Millipore, catalogue 07-634) cross-linked to 15 μl Sepharose protein A beads. Immunoprecipitations were performed for 2 h at 4°C. The immunopurified proteins were eluted from the beads while they were being shaken at 37°C, with 20 μl 2× Laemmli sample buffer without β-mercaptoethanol. After elution, sample buffer was transferred to a new vial, 1% β-mercaptoethanol was added, and samples were run on a 10% SDS polyacrylamide gel. Afterward, the gel was fixed in 25% isopropanol with 10% acetic acid, stained with Coomassie Brilliant Blue (Pageblue; Fermentas, Vilnius, Lithuania) for 1 h, and destained in water.
In-gel digestion
Gel bands were cut and processed for protein in-gel digestion as described elsewhere (Shevchenko et al., 2006). Briefly, proteins were reduced with dithiothreitol and then alkylated with iodoacetamide. Trypsin was added at a concentration of 10 ng/μl and digested overnight at 37°C. Subsequently, peptides were collected from the supernatants, and a second extraction using 10% formic acid was carried out.
Mass spectrometry analysis
Nanoflow liquid chromatography with tandem mass spectrometry was carried out by coupling an Agilent 1100 high-performance liquid chromatography system (Agilent Technologies) to an LTQ-Orbitrap XL mass spectrometer (Thermo Electron). Peptide samples were delivered to a trap column (AquaTM C18, 5 μm [Phenomenex]; 20 mm × 100 μm inner diameter, packed in-house) at 5 μl/min in 100% solvent A (0.1 M acetic acid in water). Next peptides were eluted from the trap column onto an analytical column (ReproSil-Pur C18-AQ, 3 μm, Dr. Maisch, GmbH, Ammerbuch-Entringen, Germany; 40 cm × 50 μm inner diameter, packed in-house) at ∼100 nl/min in a 90-min gradient from 0 to 40% solvent B (0.1 M acetic acid in 8:2 [vol/vol] acetonitrile/water). The eluent was sprayed using distal-coated emitter tips butt-connected to the analytical column. The mass spectrometer was operated in data-dependent mode, automatically switching between mass spectrometry and tandem mass spectrometry. Full-scan mass spectrometry spectra (from m/z 300 to m/z 1500) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000 in the linear ion trap. The two or five most-intense ions at a threshold above 5000 were selected for collision-induced fragmentation in the linear ion trap at a normalized collision energy of 35% after accumulation to a target value of 10,000.
Data analysis
Peak lists were created from raw files with MaxQuant (Cox and Mann, 2008). Peptide identification was carried out with Mascot 2.3 (Matrix Science) against a C. elegans protein database (www.wormbase.org) supplemented with all of the frequently observed contaminants in mass spectrometry (23,502 protein sequences in total). The following parameters were used: 10 ppm for precursor mass tolerance, 0.6 Da for fragment ion tolerance, and up to two missed cleavages allowed. Carbamidomethylation of cysteine was set as the fixed modification, whereas oxidation of methionine was set as the variable modification.
Y2H screening
Y2H screens were performed following a mating approach (Fromont-Racine et al., 1997). Bait clones transformed into the Y8930 yeast strain were grown overnight in YEPD (yeast extract–peptone–dextrose) medium and mixed with equal volumes of yeast containing cDNA prey libraries and fragment prey libraries, which have been described elsewhere (Boxem et al., 2008). Yeast mixtures were spun down for 30 s at 5000 rpm, resuspended in 300 μl Milli-Q (MQ), and plated on 9-cm YEPD plates. After incubation for 4 h at 30°C, yeast was washed from the plates and collected in a 2-ml tube. After another spin for 30 s at 5000 rpm, the cells were resuspended in 600 μl MQ and divided over two 15-cm −Leu/−Trp/−His plates. For estimating the number of screened diploids, a 1:10,000 dilution on a −Leu/−Trp plate was taken as a control. After 4 d of growth, resulting colonies were picked into 25 μl MQ, and 5 μl was spotted on two −Leu/−Trp/−His plates. Following 1 d of growth, one of the −Leu/−Trp/−His plates was replica-plated on −Leu/−Trp/−His + 2 mM 3-AT, −Leu/−Trp/−Ade, and −Leu/−His + 1 μg/ml cycloheximide plates. Yeast was allowed to grow for 2 d at 30°C, except for the yeast plated on −Leu/−Trp/−Ade, which was held at room temperature. After elimination of potential autoactivators (Vidalain et al., 2004), positive colonies were picked into 100 μl MQ, and 5 μl was spotted on −Leu −Trp and YEPD plates. After sufficient growth, spotted yeast colonies were lysed, and PCR was performed for prey clones; this was followed by DNA sequence analysis. Only interactions found two or more times were taken into account.
Supplementary Material
Acknowledgments
We thank H. Bringmann, P. Gönczy, E. Jorgensen, and A. Spang for strains or reagents and B. Snel for assistance in TCC-1 homology analysis. We are grateful to N. Goehring and S. Olson for advice on imaging of osmotic-sensitive embryos and A. Thomas for critically reading the manuscript. We acknowledge the Caenorhabditis Genetics Center, supported by the National Institutes of Health National Center for Research Resources, for several strains used in this study. This work is part of research program 81902016 financed by the Netherlands Organization for Scientific Research (NWO/ALW). A.A. is supported by a NWO/ALW VICI grant. The Netherlands Proteomics Center, a program embedded in the Netherlands Genomics Initiative, is kindly acknowledged for financial support.
Abbreviations used:
- AB
anterior blastomere
- DAPI
4′,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- ER
endoplasmic reticulum
- Gα
α subunit of a heterotrimeric G protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GPR-1/2
TPR-GoLoco motif proteins GPR-1 and GPR-2
- MQ
Milli-Q
- NA
numerical aperture
- PBS
phosphate-buffered saline
- RNAi
RNA interference
- TCC-1
transmembrane and coiled-coil domain protein 1
- Y2H
yeast two-hybrid
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-02-0076) on May 22, 2013.
*Present address: Proteomics Unit, Spanish National Cancer Research Center, 28029 Madrid, Spain.
REFERENCES
- Afshar K, Werner ME, Tse YC, Glotzer M, Gonczy P. Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development. 2010;137:237–247. doi: 10.1242/dev.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar K, Willard FS, Colombo K, Johnston CA, McCudden CR, Siderovski DP, Gonczy P. RIC-8 is required for GPR-1/2-dependent Gα function during asymmetric division of C. elegans embryos. Cell. 2004;119:219–230. doi: 10.1016/j.cell.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Afshar K, Willard FS, Colombo K, Siderovski DP, Gonczy P. Cortical localization of the Gα protein GPA-16 requires RIC-8 function during C. elegans asymmetric cell division. Development. 2005;132:4449–4459. doi: 10.1242/dev.02039. [DOI] [PubMed] [Google Scholar]
- Bergmann DC, Lee M, Robertson B, Tsou MF, Rose LS, Wood WB. Embryonic handedness choice in C. elegans involves the Gα protein GPA-16. Development. 2003;130:5731–5740. doi: 10.1242/dev.00839. [DOI] [PubMed] [Google Scholar]
- Boxem M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, Oegema K. Acute drug treatment in the early C. elegans embryo. PLoS One. 2011;6:e24656. doi: 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science. 2003;300:1957–1961. doi: 10.1126/science.1084146. [DOI] [PubMed] [Google Scholar]
- Couwenbergs C, Labbe JC, Goulding M, Marty T, Bowerman B, Gotta M. Heterotrimeric G protein signaling functions with dynein to promote spindle positioning in C. elegans. J Cell Biol. 2007;179:15–22. doi: 10.1083/jcb.200707085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Du Q, Macara IG. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- Ellefson ML, McNally FJ. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans. Mol Biol Cell. 2009;20:2722–2730. doi: 10.1091/mbc.E08-12-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk Green R, Lorson M, Walhout AJ, Vidal M, van den Heuvel S. Identification of critical domains and putative partners for the Caenorhabditis elegans spindle component LIN-5. Mol Genet Genomics. 2004;271:532–544. doi: 10.1007/s00438-004-1012-x. [DOI] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Gabay M, Pinter ME, Wright FA, Chan P, Murphy AJ, Valenzuela DM, Yancopoulos GD, Tall GG. Ric-8 proteins are molecular chaperones that direct nascent G protein alpha subunit membrane association. Sci Signal. 2011;4:ra79. doi: 10.1126/scisignal.2002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, Munoz J, Portegijs V, Boxem M, Grill SW, Heck AJ, van den Heuvel S. aPKC phosphorylates NuMA-related LIN-5 to position the mitotic spindle during asymmetric division. Nat Cell Biol. 2011;13:1132–1138. doi: 10.1038/ncb2315. [DOI] [PubMed] [Google Scholar]
- Galli M, van den Heuvel S. Determination of the cleavage plane in early C. elegans embryos. Annu Rev Genet. 2008;42:389–411. doi: 10.1146/annurev.genet.40.110405.090523. [DOI] [PubMed] [Google Scholar]
- Gassmann R, Holland AJ, Varma D, Wan X, Civril F, Cleveland DW, Oegema K, Salmon ED, Desai A. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–971. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Hoege C, Grill SW, Hyman AA. PAR proteins diffuse freely across the anterior-posterior boundary in polarized C. elegans embryos. J Cell Biol. 2011;193:583–594. doi: 10.1083/jcb.201011094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;5:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Pichler S, Kirkham M, Hyman AA. Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol. 1999;9:135–150. doi: 10.1083/jcb.147.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Dong Y, Peterson YK, Lanier SM, Ahringer J. Asymmetrically distributed C. elegans homologs of AGS3/PINS control spindle position in the early embryo. Curr Biol. 2003;13:1029–1037. doi: 10.1016/s0960-9822(03)00371-3. [DOI] [PubMed] [Google Scholar]
- Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kimura A. Intracellular organelles mediate cytoplasmic pulling force for centrosome centration in the Caenorhabditis elegans early embryo. Proc Natl Acad Sci USA. 2011;108:137–142. doi: 10.1073/pnas.1013275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;4:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson MA, Horvitz HR, van den Heuvel S. LIN-5 is a novel component of the spindle apparatus required for chromosome segregation and cleavage plane specification in Caenorhabditis elegans. J Cell Biol. 2000;148:73–86. doi: 10.1083/jcb.148.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrari Y, Crouthamel M, Irannejad R, Wedegaertner PB. Assembly and trafficking of heterotrimeric G proteins. Biochemistry. 2007;46:7665–7677. doi: 10.1021/bi700338m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, Martin JL, Ellefson M, McNally FJ. Kinesin-dependent transport results in polarized migration of the nucleus in oocytes and inward movement of yolk granules in meiotic embryos. Dev Biol. 2010;339:126–140. doi: 10.1016/j.ydbio.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Ahearn I, Bergo M, Young S, Philips M. Membrane trafficking of heterotrimeric G proteins via the endoplasmic reticulum and Golgi. Mol Biol Cell. 2002;13:3294–3302. doi: 10.1091/mbc.E02-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nakayama Y, Shivas JM, Poole DS, Squirrell JM, Kulkoski JM, Schleede JB, Skop AR. Dynamin participates in the maintenance of anterior polarity in the Caenorhabditis elegans embryo. Dev Cell. 2009;16:889–900. doi: 10.1016/j.devcel.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc T, Afshar K, Gonczy P. Coupling of cortical dynein and Gα proteins mediates spindle positioning in Caenorhabditis elegans. Nat Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- Panbianco C, Weinkove D, Zanin E, Jones D, Divecha N, Gotta M, Ahringer J. A casein kinase 1 and PAR proteins regulate asymmetry of a PIP(2) synthesis enzyme for asymmetric spindle positioning. Dev Cell. 2008;15:198–208. doi: 10.1016/j.devcel.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park DH, Rose LS. Dynamic localization of LIN-5 and GPR-1/2 to cortical force generation domains during spindle positioning. Dev Biol. 2008;315:42–54. doi: 10.1016/j.ydbio.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell. 2005;16:2139–2153. doi: 10.1091/mbc.E04-08-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Pecreaux J, Goehring NW, Khairy K, Stelzer EH, Hyman AA, Howard J. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS One. 2010;5:e12301. doi: 10.1371/journal.pone.0012301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek M, Thomas JH. Calcium/calmodulin-dependent protein kinase II regulates Caenorhabditis elegans locomotion in concert with a Go/Gq signaling network. Genetics. 2000;156:1069–1082. doi: 10.1093/genetics/156.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–919. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Bowerman B. Myosin and the PAR proteins polarize microfilament-dependent forces that shape and position mitotic spindles in Caenorhabditis elegans. J Cell Biol. 2003;161:21–26. doi: 10.1083/jcb.200210171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Shivas JM, Skop AR. Arp2/3 mediates early endosome dynamics necessary for the maintenance of PAR asymmetry in Caenorhabditis elegans. Mol Biol Cell. 2012;23:1917–1927. doi: 10.1091/mbc.E12-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode Caenorhabditis elegans. J Cell Biol. 1986;103:2241–2252. doi: 10.1083/jcb.103.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagawa A, Rappleye CA, Aroian RV. Pod-2, along with pod-1, defines a new class of genes required for polarity in the early Caenorhabditis elegans embryo. Dev Biol. 2001;233:412–424. doi: 10.1006/dbio.2001.0234. [DOI] [PubMed] [Google Scholar]
- Thyagarajan K, Afshar K, Gonczy P. Polarity mediates asymmetric trafficking of the Gβ heterotrimeric G-protein subunit GPB-1 in C. elegans embryos. Development. 2011;138:2773–2782. doi: 10.1242/dev.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Hayashi A, DeBella LR, McGrath G, Rose LS. LET-99 determines spindle position and is asymmetrically enriched in response to PAR polarity cues in C. elegans embryos. Development. 2002;129:4469–4481. doi: 10.1242/dev.129.19.4469. [DOI] [PubMed] [Google Scholar]
- van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Gα. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- Vidalain PO, Boxem M, Ge H, Li S, Vidal M. Increasing specificity in high-throughput yeast two-hybrid experiments. Methods. 2004;32:363–370. doi: 10.1016/j.ymeth.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Welte MA. Bidirectional transport along microtubules. Curr Biol. 2004;14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Werts AD, Roh-Johnson M, Goldstein B. Dynamic localization of C. elegans TPR-GoLoco proteins mediates mitotic spindle orientation by extrinsic signaling. Development. 2011;138:4411–4422. doi: 10.1242/dev.070979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MJ, Bola B, Brownhill K, Yang YC, Levakova V, Allan VJ. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J Cell Sci. 2009;122:1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–457. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Skop AR, White JG. Src and Wnt signaling regulate dynactin accumulation to the P2-EMS cell border in C. elegans embryos. J Cell Sci. 2008;121:155–161. doi: 10.1242/jcs.015966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.