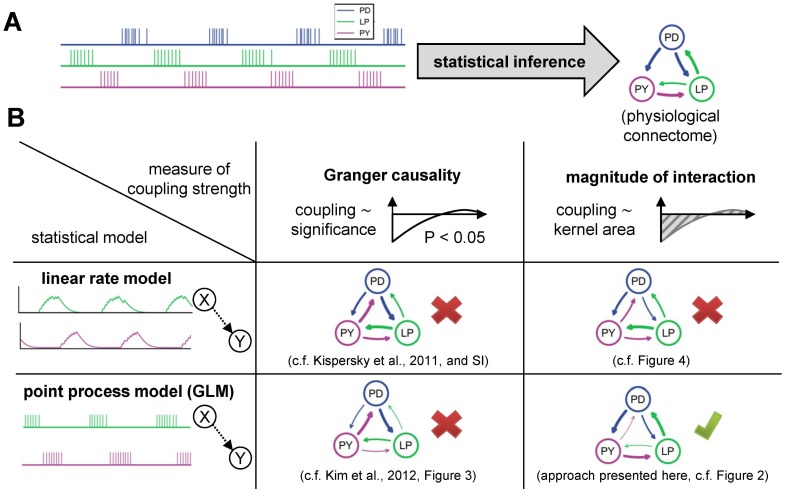
Figure 1. Inferring network connectivity of the pyloric circuit of the crab stomatogastric ganglion (STG) based on extracellular spike train recordings.
A, Statistical models fitted on spike train activity (left) can be used to infer the effective coupling. The effective coupling should match the physiologically known diagram of the pyloric circuit (right). All synaptic couplings in the pyloric circuit are inhibitory. B, Comparison of algorithms for network inference. Neural activity can be either described in a firing rate model, e.g., in classical time series analysis, or using a point process model or generalized linear model (GLM). For both models, couplings are introduced as interaction kernels between the stochastic processes. The strength of the interaction can be either quantified through its statistical significance, i.e., a Granger causality-type measure, or through the magnitude of the interaction, as measured by the net area under the interaction kernel. Only the combination of a point-process-based generalized linear model with the definition of coupling strength as the magnitude of the interaction is able to recover a connectivity that is consistent with physiology (lower right). All other combinations of models and measures infer inaccurate connectivity patterns.