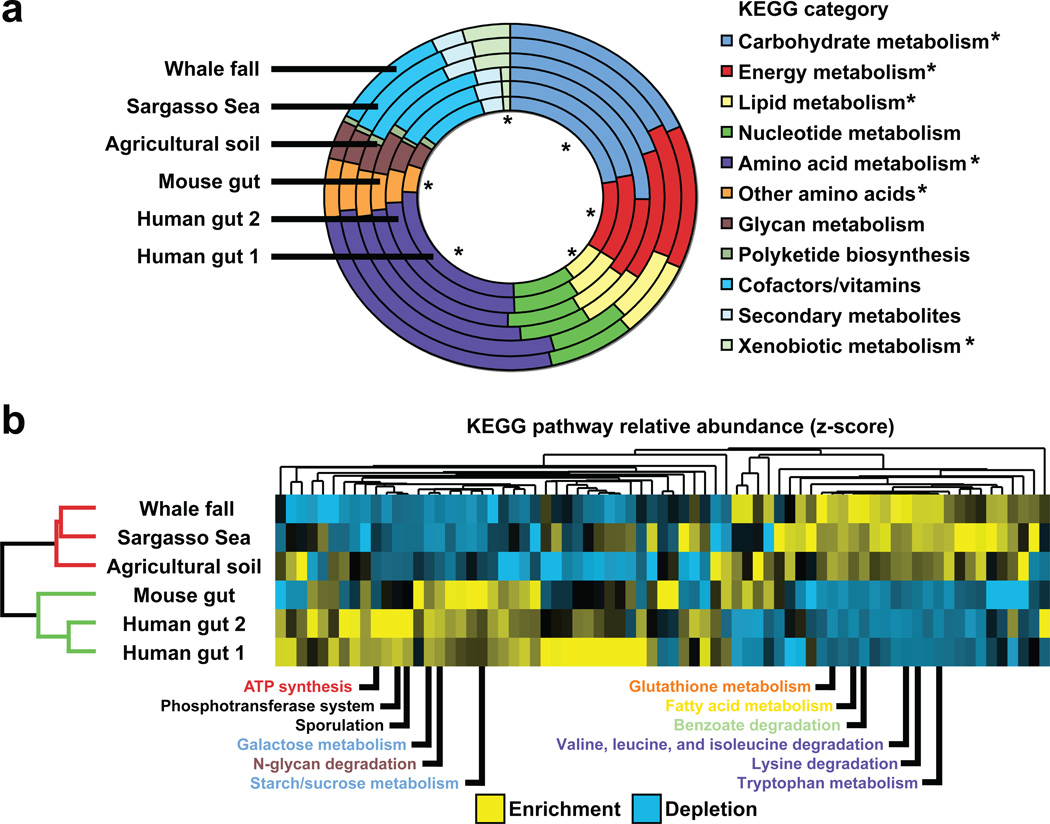
Figure 2. Functional comparisons of the gut microbiome versus other sequenced microbiomes1,19–21.
(A) Relative abundance of predicted genes assigned to KEGG categories for metabolism. Analyses were performed on the combined mouse gut dataset (n=5 animals), both human gut datasets, and three ‘environmental’ datasets: the combined Whale fall dataset (n=3 samples), agricultural soil, and the combined Sargasso Sea dataset (n=7 samples). Forward sequencing reads were culled from each dataset and mapped onto reference microbial and eukaryotic genomes from the KEGG database22 (version 40; BLASTX best-blast-hit e-value<10−5). Asterisks indicate categories that are significantly enriched or depleted in the combined gut dataset versus the combined environmental dataset (the distribution of ~15,000 KEGG pathway assignments across each of the six datasets was used to construct two combined datasets of ~45,000 KEGG pathway assignments each; Χ2 test using the Bonferroni correction for multiple hypotheses, p<10−4). (B) Hierarchical clustering based on the relative abundance of predicted proteins assigned to KEGG pathways reveals specific differences between gut (green) and environmental (red) microbiomes. The relative abundance of pathways that exceeded a threshold of >0.6% (assignments to a given pathway divided by assignments to all pathways) in at least two environments was transformed into a z-score (yellow=enrichment; blue=depletion), and clustered by environments and pathways20 using a Euclidean distance metric (Cluster 3.040). The results were visualized in Treeview41. Environmental clustering was consistent using multiple distance metrics, including Pearson Correlation (centered/uncentered), Spearman Rank Correlation, Kendall’s tau, and City-block distance. The twelve most discriminating KEGG pathways are listed (based on the ratio of average gut relative abudance versus average environmental relative abundance). Metabolic pathway names are colored based on KEGG category [pathways not colored include sporulation (cell growth/death) and phosphotranferase system (membrane transport)]. The gut microbiome is enriched for pathways involved in importing and degrading polysaccharides and simple sugars (‘starch/sucrose metabolism’, ‘galactose metabolism’, ‘N-glycan degradation’, and ‘phosphotransferase system’). The gut microbiome is also enriched for genes involved in ‘sporulation’, reflecting the high relative abundance of Gram-positive Firmicutes.