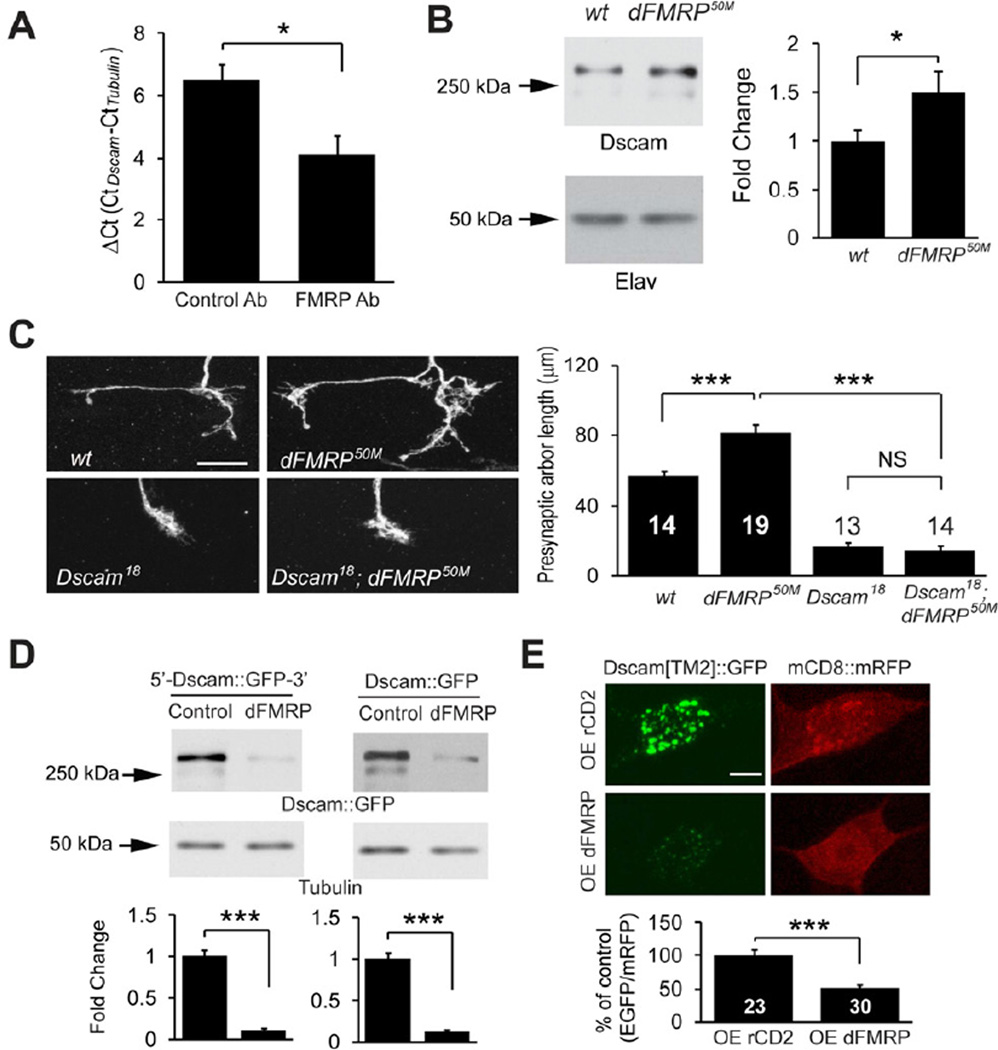
Figure 6. FMRP Suppresses Dscam Expression to Restrict Presynaptic Arbor Growth.
(A) dFMRP associates with Dscam mRNA in vivo. RNA-immunoprecipitation, followed by reverse transcription and real-time PCR, was done using larval brain lysates (n = 3). The difference in DCt between control and dFMRP-immunoprecipitates reflects a 5.8-fold binding of Dscam mRNA to FMRP. (B) Western analysis of Dscam expression in the brains of wild-type (w1118) and dFMRP50M third instar larvae. The intensities of Dscam bands were normalized to those of the neuron-specific protein Elav and presented as fold change (n = 13). (C) Dscam is required by dFMRP to restrict presynaptic arbor growth. Images show the presynaptic arbors of ddaC MARCM clones of wt, dFMRP50M, sDscam18, and Dscam18/dFMRP50M double mutant (Dscam18; dFMRP50M) neurons. Scale bar: 10 µm. Right: Quantification of presynaptic arbor length for each condition. Sample numbers are shown in or above the bars. (D) dFMRP regulates Dscam expression through the coding region of Dscam in S2 cells. Shown are Western blots of lysates of cultured S2 cells expressing Dscam[TM2] ∷ GFP with Dscam 5’ and 3’UTR (5’-Dscam ∷ GFP-3’) or with SV40 3’UTR (Dscam ∷ GFP) in the presence of a dFMRP-expression construct (dFMRP) or the empty vector (Control). Dscam[TM2] ∷ GFP levels were normalized to tubulin levels and presented as fold change (n = 4). (E) dFMRP suppresses Dscam expression through the Dscam coding region in vivo. Dscam[TM2] ∷ GFP and mCD8 ∷ RFP were expressed in C4da neurons using Gal44–77, along with either rCD2 (control) or dFMRP (OE dFMRP). Dscam[TM2] ∷ GFP levels in ddaC cell bodies were normalized to mCD8 ∷ mRFP levels and presented as % of control (right panel). Also see Figure S4.