Abstract
The ErbB2/3 heterodimer plays a critical role in breast cancer progression and in the development of endocrine resistance. EBP1, an ErbB3 binding protein, inhibits HRG-stimulated breast cancer growth, decreases ErbB2 protein levels and contributes to tamoxifen sensitivity. We report here that ectopic expression of EBP1 in Estrogen Receptor (ER) positive breast cancers that express ErbB2 at both high and low levels decreased ErbB2 protein levels. ErbB2 protein expression was also increased in mammary glands of Ebp1 knock out mice. To define the mechanism of ErbB2 down regulation, we examined the effects of EBP1 on ErbB2 mRNA levels, transcription of the ErbB2 gene and ErbB2 protein stability. We found that ectopic expression of EBP1 decreased steady state levels of endogenous ErbB2 mRNA in all cell lines tested. EBP1 overexpression decreased the activity of an ErbB2 promoter reporter in cells which overxpress ErbB2. However, reporter activity was unchanged or increased in cells which express low endogenous levels of ErbB2. We also found that ectopic expression of EBP1 accelerated ErbB2 protein degradation and enhanced ErbB2 ubiquitination in cells which express both low and high levels of ErbB2. Treatment with proteasome inhibitors prevented this decrease in ErbB2 protein levels. Ablation of EBP1 expression led to tamoxifen resistance that was abrogated by inhibition of ErbB2 activity. These results suggest that EBP1 inhibits expression of ErbB2 protein levels by multiple mechanisms and that EBP1’s effects on tamoxifen sensitivity are mediated in part by its ability to modulate ErbB2 levels.
Keywords: EBP1, ErbB2, tamoxifen, breast cancer
INTRODUCTION
A wealth of clinical data has demonstrated the aberrant expression of ErbB family members in breast cancer [1] [2]. The ErbB2 gene is amplified in 20-30% of breast carcinomas contributing to more aggressive disease [3]. The overexpression of ErbB2 has been successfully exploited therapeutically by use of the monoclonal antibody Trastuzumab and by tyrosine kinase inhibitors. ErbB3 is also overexpressed in many breast tumors [4] and coexpression of ErbB2 and ErbB3 is significantly associated with decreased survival [5]. Oncogenic signaling through ErbB3 can abrogate the effectiveness of ErbB directed therapies [6]. The ErbB2/ErbB3 receptor pair forms the most potent mitogenic receptor complex in vitro [7] and is key to the proliferation of human breast cancer cells that express these receptors [8].
Endocrine resistance is often associated with enhanced expression of members of the ErbB receptor family, especially ErbB2. Multiple clinical studies indicate that ErbB2 expression portends a poorer prognosis with tamoxifen therapy [9]. This is important as approximately half of breast cancers that overexpress ErbB2 also express hormone receptors [10]. It has been demonstrated in both cell culture and animal models for many years that the enhanced expression of ErbB2 leads to the ability of cells to bypass normal endocrine responsiveness [11] [12].
The biological activity and expression of ErB2 and ErbB3 are regulated by a host of interacting proteins that may be potential targets for development of new therapies. Our laboratory has been interested in proteins that regulate ErbB3, as the ErbB3 receptor has impaired tyrosine kinase activity [13, 14], necessitating its interactions with other proteins to exert its biological effects. An ErbB3 binding protein (EBP1) was isolated in our laboratory during a yeast two-hybrid screen for ErbB3 interacting proteins [15]. Overexpression of EBP1 inhibits growth of ErbB2/3 expressing cell lines. Ectopic expression of EBP1 promotes G2/M cell cycle arrest and cellular differentiation [16]. Overexpression of EBP1 inhibits the transcription of reporter genes controlled by Cyclin D1, Cyclin E and c-myc promoters and the transcription of endogenous E2F1 and c-myc genes via its binding to an E2F1 consensus element [17-19]. The ability of EBP1 to repress transcription requires its interaction with histone deacetylase 2 (HDAC2), Rb and Sin3A [18, 19] [20].
Our previous work has demonstrated that ectopic expression of EBP1 inhibits growth of MCF-7 and AU565 breast cancer cell lines in response to HRG and interferes with HRG induced growth signals, such as the activation of AKT. In addition, ErbB2 protein levels are decreased in MCF-7 and AU565 cells transfected with EBP1 [21]. In the current study, we were interested in determining the mechanism of ErbB2 downregulation in ER+ breast cancer cells. We found that EBP1 decreased steady state levels of ErbB2 mRNA in all cell lines tested. However, ErbB2 promoter activity was decreased only in cells which overexpress ErbB2. EBP1 decreased ErbB2 protein stability in cells which express both high and low levels of ErbB2 via a proteasome mediated pathway. We also found that tamoxifen resistance induced by ablation of EBP1 expression was abrogated by inhibition of ErbB2 activity.
MATERIALS AND METHODS
Cell Culture
MCF-7, T47D and BT474 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained at 37°C in a humidified atmosphere of 5% CO2 in air in RPMI 1640 (Biofluids, Rockville, MD) and 10% FBS (Sigma, St. Louis, MO).
Reagents
Heregulin β1 (HRGβ1) was obtained from R & D Systems Inc. (Minneapolis, MN), EGF and 4-hydroxy-tamoxifen (OHT) from Sigma, and Geneticin (G418) from Invitrogen (Carlsbad, CA). The proteasome inhibitor ALLnL was from Sigma and MG132 from Calbiochem (San Diego, CA).
Plasmids
A full-length EBP1 cDNA(GenBank NM006191) was generated by PCR with specific reverse and forward primers containing EcoRI and BamHI restriction sites using a pcDNA-EBP1 vector as a template [17]. This cDNA includes all 3 possible translation initiation sites of EBP1 and encodes the largest form of the protein. The EGFP-EBP1 plasmid was constructed by cloning full length EBP1 into the BamHI/ EcoRI sites of the EGFP-C1 vector (Clontech, Palo Alto CA) and the EcoRI/BamHI sites of the CMV10 vector (Sigma) respectively. The pcDNA3 ErbB2 expression plasmid was a generous gift of Dr. Yossi Yarden.
Creation of stably transfected cell lines
To establish EBP1 overexpressing stable transfectants, subconfluent cells in 100-mm tissue culture dishes were transfected with 10μg of EGFP, or EGFP- EBP1 expression plasmids using Fugene-6 (Roche, Indianapolis, IN) according to the manufacturer’s protocol. Cells were selected in G418 (500 μg/ml) for 4 weeks and mass cultures obtained.
To generate EBP1 silenced cell lines, cells were seeded into 96 well plates and transduced with lentiviral particles (MOI 25) with shRNA targeted to NT 302-322 of the PA2G4 (EBP1) (Genbank NM_006191.1) gene (Mission shRNA, Sigma) according to the manufacturer’s protocol. Cell lines were selected in 2 μg/ml of puromycin and surviving colonies of cells expanded as mass cultures. Four independent transductions of this lentiviral construct were performed and designated Clones 1-4. The control was an EBP1 directed lentivirus (NT 776-793) that was previously demonstrated not to change EBP1 expression [22].
Western blot Analysis
Total cell extracts were prepared by direct lysis of cells with lysis buffer [50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 250 mM NaCl, 1% Triton X-100, 0.5 mM DTT, and 1× Complete™ protease inhibitor]. Protein concentrations were measured using a detergent compatible kit (Bio-Rad, Richmond, CA). Proteins (30 μg per well) were resolved by SDS-PAGE, transferred to PVDF membranes, and immunoblotted as described [15]. The EBP1 antibody was from Upstate (Lake Placid, NY), the anti –actin and GAPDH antibodies from Sigma, the ubiquitin antibodies were from Cell Signaling (Danvers, MA). Images were quantified using IMAGE-J software (NIH). Where indicated, blots were stripped in Restore Western blot Stripping Buffer™ (Pierce, Rockford, IL) as directed by the manufacturer and reprobed.
Immunohistochemical analysis
Mammary glands (#4) were excised and fixed in 10% buffered neutral formalin. Sections of formalin-fixed, paraffin-embedded tissues were cut to 5 μm. Slides were stained with rabbit ErbB2 and EBP1 antibodies (both from Upstate) diluted 1:100 using the standard avidin-biotin method (Vecta-Stain Kit,Vector Labs, Burlinghame, CA) with Harris hematoxylin as a counterstain [23].
Cell Growth Assays
For studies assessing the effect of tamoxifen on cell growth, cells (5× 103) were plated in 96 well plates in complete media. After a 24-hour attachment period, the medium was replaced with complete medium containing hydroxy-tamoxifen (OHT) at the indicated concentrations. Relative live cell numbers were determined at Day 4 using a Promega Proliferation Reagent (Promega, Madison,WI) as per manufacturer’s instructions with absorbance being read at 490nm using a Dynex plate reader.
RNA Isolation and PCR analysis
RNA was extracted from whole cell lysates, DNAse treated and converted into cDNA using an AMV reverse-transcription system (Promega) in the presence of random hexamers (Invitrogen).The cDNA was used for conventional PCR or quantitative real-time PCR (RT-qPCR) with specific gene primers as follows: ErB2 forward 5′- gggaagaatggggtcgtcaaa 3′ or agccgcgagcacccaagt and reverse 5′- ctcctccctggggtgtcaagt or ttggtgggcaggtaggtgagtt GAPDH (NM_002046): forward 5′- ccacccatggcaaattcc -3′ and Reverse 5′- tcgctcctggaagatggtg. An MYIQ real-time PCR detection system and SYBR green PCR mix (BioRad) were used to carry out the real-time PCR. The relative quantitation of targeted genes was determined by the comparative ΔΔCt (threshold) method using GAPDH as an internal control [24]. All data were analyzed from three independent experiments and statistical significance was validated by Student’s t-test.
Dual Luciferase Assay
5 × 104 cells/well were transfected with 0.5μg of GFP or GFP-EBP1, 0.5μg of ErbB2-luc (a firefly luciferase reporter gene under the control of the −500 to +1 region of the ErbB2 promoter [25] and 5ng of the pRL-TK vector (a Renilla luciferase reporter gene under the control of the thymidine kinase promoter) using Fugene 6. Forty-eight hours after transfection, cells were lysed and luciferase activity determined using a dual-luciferase reporter assay (Promega). The activities of Renilla luciferase were used to normalize any variations in transfection efficiency.
Statistical Analysis
Data were analyzed using a two-tailed Students t-test and a p < 0.05 was deemed statistically significant.
RESULTS
Modulation of EBP1 expression affects ErbB2 protein levels
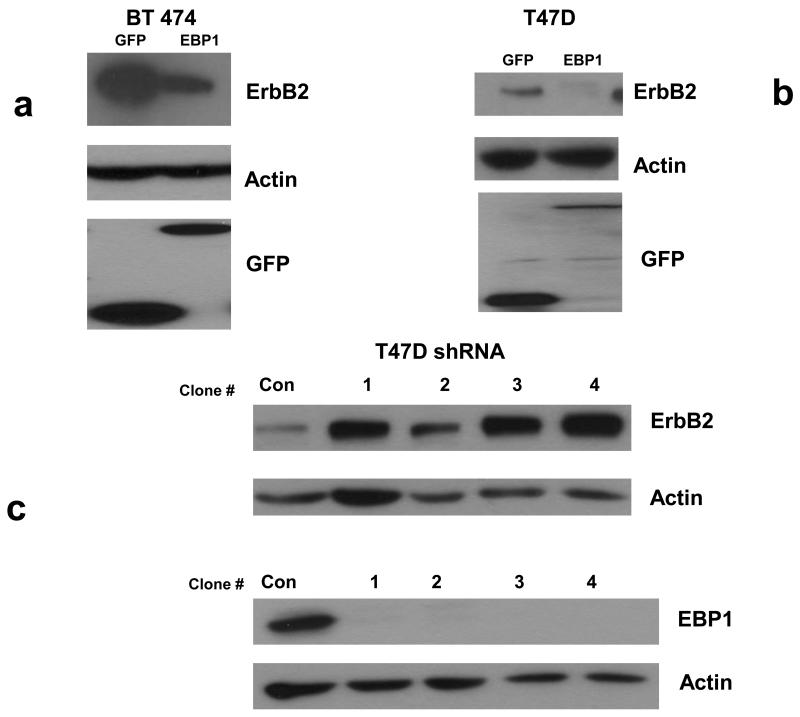
Our previous published results indicated that ectopic expression of EBP1 decreased protein levels of ErbB2 in MCF-7 (ER+) and AU565 (ER −) cells [21]. Overexpression of ErbB2 is a major problem in development of endocrine resistance in ER+ cell lines [9] and devising new ways to decrease ErbB2 levels are important for future therapeutic development. We were therefore interested in assessing the effects of EBP1 in ER+ cells lines that express ErbB2 at both high and low levels. We overexpressed EBP1 in ER+ ErbB2 high BT474 cells and ER+, ErbB2 low T47D cells. Ectopic expression of EBP1 decreased ErbB2 levels three fold in BT474 cells and five- fold in T47D cells as determined by densitometric analysis (Fig. 1a,b). However, the levels of ErbB2 in BT474 EBP1 transfectants were still much greater than those observed in control T47D cells. Elimination of EBP1 expression by shRNA in T47D cells enhanced ErbB2 protein levels four fold (Fig. 1c) as we previously described for MCF-7 cells [21].
Fig. 1. Effect of modulation of EBP1 expression on ErbB2 protein levels.
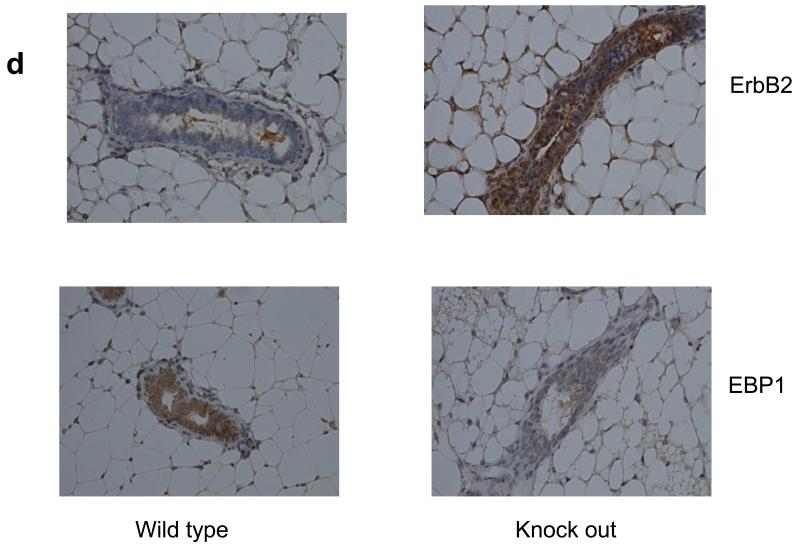
a,b) Lysates of logarithmically growing BT474 or T47D vector control cells (GFP) or EBP1 transfected cells (EBP1) were analyzed by Western blotting with antibodies for ErbB2, Actin or GFP as indicated. Data are representative of 3 experiments. c) T47D cells were transduced with Control (Con) or EBP1 (1-4) directed lentiviruses and ErbB2, EBP1 or actin levels measured by Western blotting. Clones 1-4 are directed against the same EBP1 NT sequence, but represent independent experiments. d. Number 4 mammary glands were isolated from female 10 week old wild type (3) or Ebp1 knock out mice (3). Immunohistochemical analysis was performed for ErbB2 or EBP1 as indicated. A representative field is shown.
To further determine the physiological effect of Ebp1 on endogenous ErbB2 levels, we examined expression of ErbB2 in adult mammary epithelium in a knock out mouse we recently developed [26]. We found that in mammary epithelial cells, ErbB2 expression was increased in mammary tissue of Ebp1−/− 10 week old female mice (Fig. 1d).
Steady state mRNA levels are decreased after EBP1 transfection
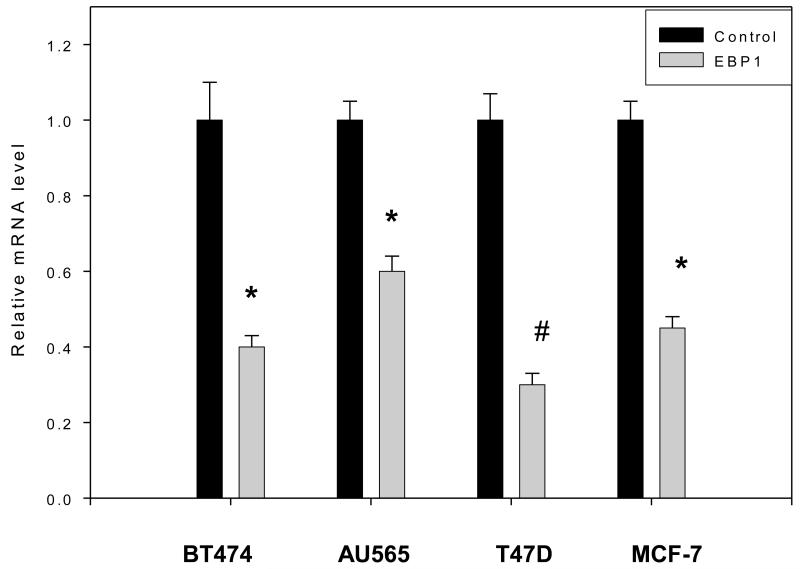
To determine a mechanism for the decreased protein levels of ErbB2, we first examined steady state ErbB2 mRNA levels in EBP1 transfected and control cell lines which express high (BT474, AU565) or low (MCF-7, T47D) levels of ErbB2. The expression of the EBP1-GFP transgene is shown in Figure 1a,b for T47D and BT474 cells lines. The expression of the FLAG-tagged EBP1 in the AU565 and MCF-7 EBP1 transfectants has previously been published [21]. We found by RT-qPCR that ectopic expression of EBP1 led to statistically significant decreases in steady state ErbB2 mRNA levels in all cell lines tested (Fig. 2). ErbB2 mRNA levels were decreased about 50% in BT474, AU565 and MCF-7 cells and 66% in T47D cells. However, the levels of ErbB2 mRNA found in BT474 and AU565 cells stably overexpressing EBP1 were significantly higher than those found in control T47D and MCF-7 cells. Thus, while EBP1 overexpression lowers ErbB2 mRNA levels in cells expressing high levels of ErbB2, mRNA levels do not reach those observed in cells with normally low endogenous levels.
Fig. 2. Steady state levels of ErbB2 mRNA in vector control and EBP1 transfected cells.
The cells lines indicated were stably transfected with EBP1 (see Fig. 1 and [21]. Total RNA (triplicate wells) was then extracted to detect the steady-state levels of ErbB2 and GAPDH mRNAs by quantitative RT-PCR. The mRNA level observed in control BT474 cells was set at 1. All values are presented in relation to this standard. Bars represent means ± SEM. The graph is representative of one of three independent experiments. *P<0.05 compared with control vector; #P<.01 compared with control vector.
Ectopic expression of EBP1 affects the activity of the ErbB2 promoter
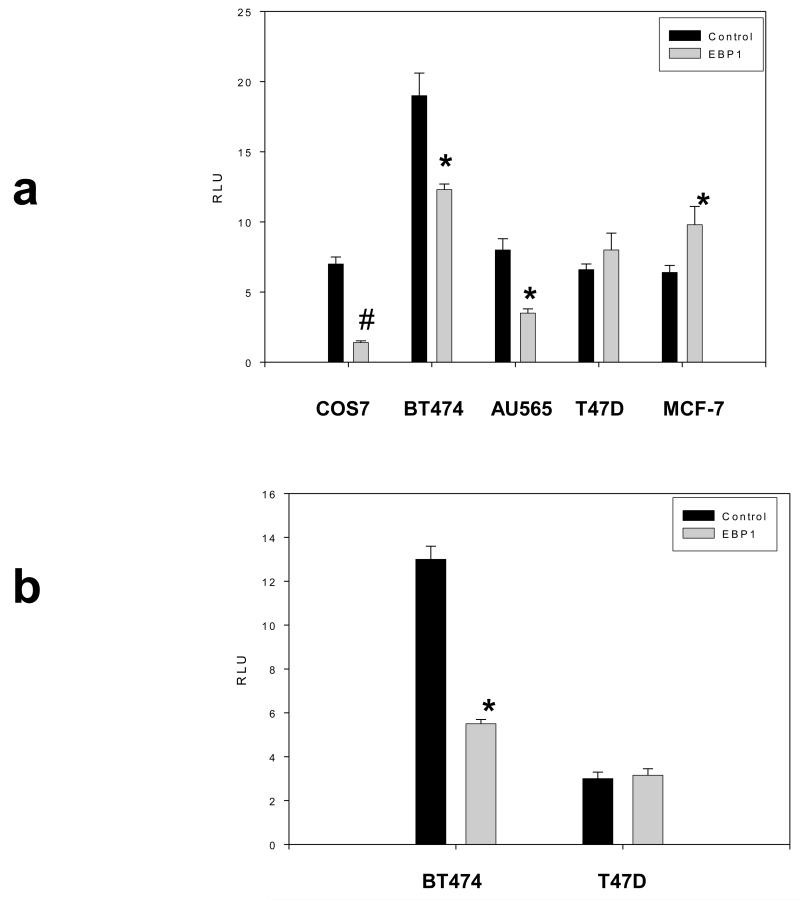
We next determined if EBP1 could affect ErbB2 transcription using an ErbB2 promoter – luciferase reporter that encodes the 500 bp proximal promoter upstream from the transcription start site. Transient transfection of EBP1 into COS7, BT474, and AU565 cells led to significant decreases in reporter activity (Fig.3a). However, the level of reporter activity in BT474 EBP1 transfectants was higher than that observed in T47D controls. In contrast, the activity of the reporter was not significantly changed in T47D and significantly increased in MCF-7 cells (Fig. 3a). In EBP1 stable BT474 transfectants,a statistically significant 58% decrease in luciferase activity was observed as compared to vector controls (Fig. 3b). No changes in reporter activity were observed in T47D stable EBP1 transfectants (Fig. 3b).
Fig. 3. Effect of Ebp1 on ErbB2 promoter activity.
a) The cell lines indicated were transiently co-transfected with ErbB2-luc, pRL-TK and GFP (Control) or GFP-EBP1. After 48hrs, cells were lysed and relative luciferase units were determined as described in the Material and Methods. The data are expressed as Relative Light Units (RLU) which is the ratio of ErbB2-luc RLU: pRL-TK RLU for each sample. Each bar represents the mean ± S.D. of 8 wells. The figure is representative of 3 independent experiments. b) Cells lines stably transfected with EBP1 or GFP (control) were transiently transfected with ErbB2-luc and pRL-TK. Luciferase activity was determined as described in A. *P<0.05 compared with control vector; #P<.01 compared with control vector.
Effects of EBP1 on ErbB2 protein stability
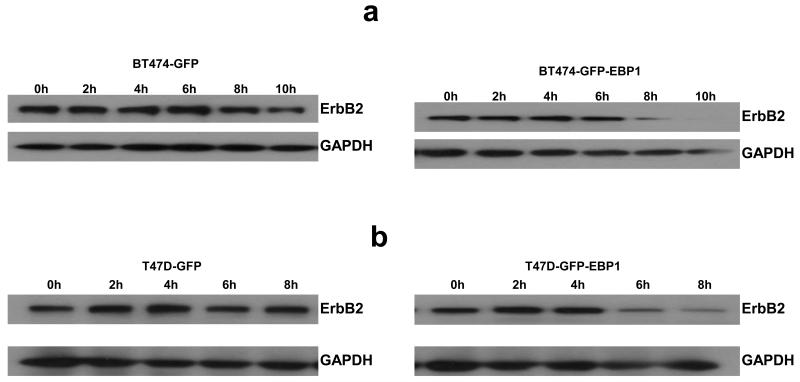
As ErbB2 protein levels were decreased more than mRNA in both BT474 and T47D EBP1 stable transfectants, we next examined ErbB2 protein stability. ErbB2 protein levels have been demonstrated to be regulated by proteasome mediated degradation [27]. We examined ErbB2 levels after inhibition of protein biosynthesis with cycloheximide (20 μg/ml). We found that in BT474 cells the expression of ErbB2 protein was significantly lower at 8 hours after cycloheximide treatment as compared to vector controls (Fig. 4a). In T47D cells, ErbB2 protein levels were also lower after 8 hours of cycloheximide treatment in EBP1 transfected cells (Fig. 4b) as compared to vector controls.
Fig. 4. Decreased stability of ErbB2 protein after ectopic expression of EBP1.
Control GFP or EBP1 stably transfected BT474 (a) or T47D cells (b) were treated with cycloheximide. Cells lysates were obtained at the indicated time points and analyzed by Western blotting with ErbB2 or GAPDH antibodies as indicated.
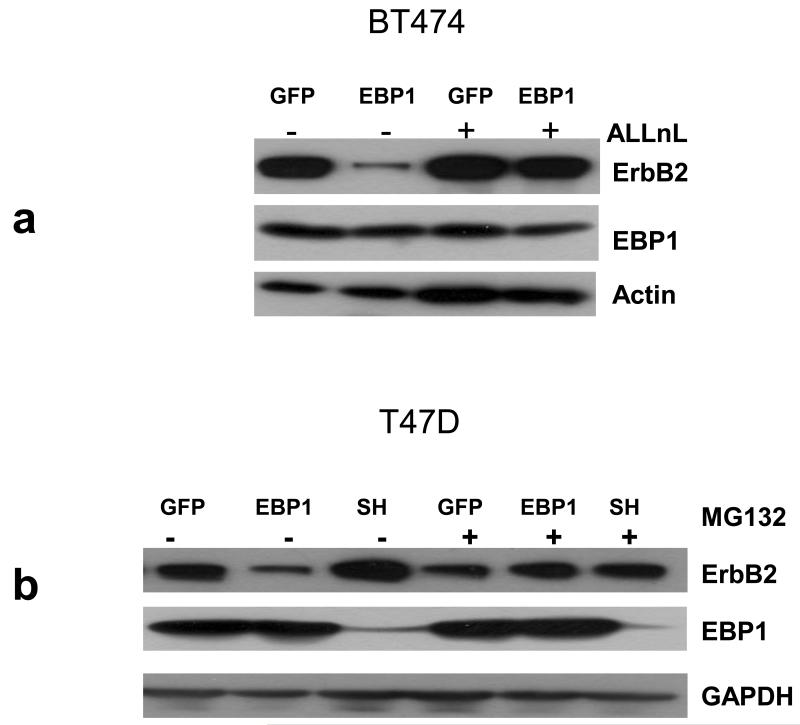
We next determined if the increased ErbB2 degradation observed after ectopic expression of EBP1 could be rescued by proteasome inhibition. We used the proteasome inhibitors MG132 and ALLnL to stabilize ubiquitinated ErbB2 in BT474 and T47D cells in which EBP1 had been overexpressed. As expected, ErbB2 levels in the absence of the inhibitor were decreased in both BT474 and T47D EBP1 transfectants as compared to controls (Figs. 5a,b). ErbB2 levels were increased when EBP1 was ablated in T47D cells (Fig. 5B). Addition of ALLnL was able to overcome the decrease in ErbB2 protein observed in BT474 EBP1 transfectants. However, MG132 was not able to restore ErbB2 levels (data not shown). This is of interest as ALLnL inhibits the activity of both the proteasome and calpain I [27]. In T47D EBP1 transfectants both MG132 (Fig. 5b) and ALLnL (data not shown) resulted in restoration of ErbB2 levels. In EBP1 silenced cells, ErbB2 levels were slightly decreased in the presence of the proteasome inhibitor (Fig. 5b). EBP1 itself has been demonstrated to be a target of ubiquitination. Two isoforms of EBP1 have been reported: a p42 pro-apoptotic form and a p48 anti-apoptotic form. The p42 isoform appears to be preferentially degraded in cancer cells [28]. However, endogenous EBP1 levels were not increased by proteasome inhibitors in either cell line at this time point. Only the 48kD form of EBP1 was observed (Fig. 5a, b). The GFP-EBP1 fusion protein continued to be overexpressed at about the same level (data not shown).
Fig. 5. EBP1 induced degradation of ErbB2 can be resuced by proteasome inhibition.
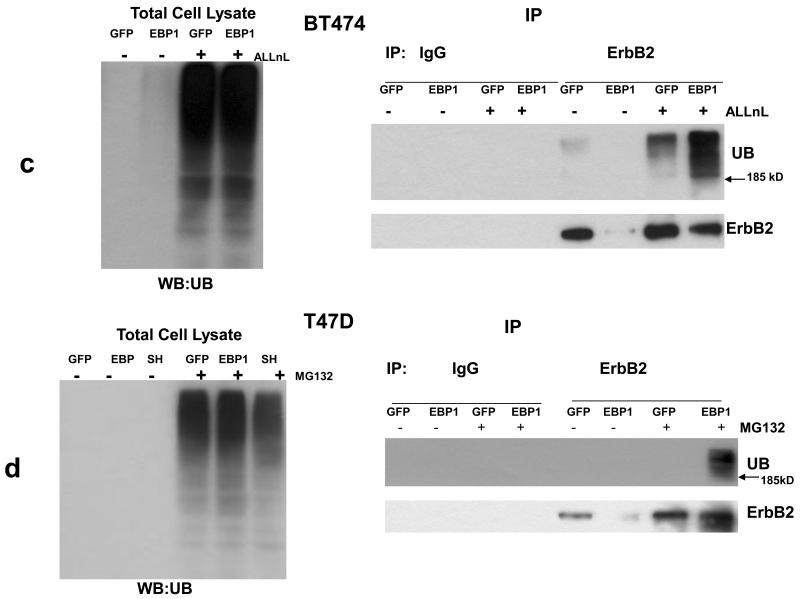
a,b) BT474 GFP control and EBP1 transfected cells(a) and T4D control (GFP) cells and cells in which EBP1 was transfected (EBP1) or ablated (SH) (b) were treated with the indicated proteasome inhibitors for 6 hours prior to lysis. Cells lysates were analyzed by Western blotting with antibodies against ErbB2, EBP1 or actin or GAPDH as indicated. c,d BT474 GFP control and EBP1 transfected cells and T4D control (GFP) cells and cells in which EBP1 was transfected (EBP1) or ablated (SH) (d) were treated with the indicated proteasome inhibitors for 6 hours prior to lysis. (Left panels) Cell lysates were resolved by SDS-PAGE and analyzed by Western blotting using an antiubiquitin antibody. (Right Panels) ErbB2 was immunoprecipitated from these cell lysates with IgG or anti-ErbB2 antibodies. Precipitated proteins were analyzed by Western blotting with antibodies to Ubiquitin or ErbB2 as indicated. Representative of three independent experiments.
Since ErbB2 is degraded through a mechanism that involves its polyubiquitination [29], we tested if ectopic expression of EBP1 could elevate ubiquitination of ErbB2. Examination of whole cell lysates revealed that treatment with proteasome inhibitors increased ubiquitination as expected (Fig.5c,d left panels). ErbB2 was next immunoprecipitated from lysates of cells treated with proteasome inhibitors for 6 hours and proteins analyzed by western blotting with anti-ubiquitin and ErbB2 antibodies. Ubiquitination of ErbB2 was enhanced by treatment with proteasome inhibitors as expected[27] in BT474 Cells (Fig. 5c, right panel). In contrast, ubiquitinated ErbB2 was not observed in the presence of MG132 in T47D GFP controls. This suggests that there is a very low steady-state level of ubiquitination of ErbB2 in control T47D cells. ErbB2 ubiquitination was increased in EBP1 transfected cells as compared to vector controls for both cell lines (Fig. 5, C,D right panels).
EBP1 affects the response to tamoxifen through an ErB2-mediated pathway
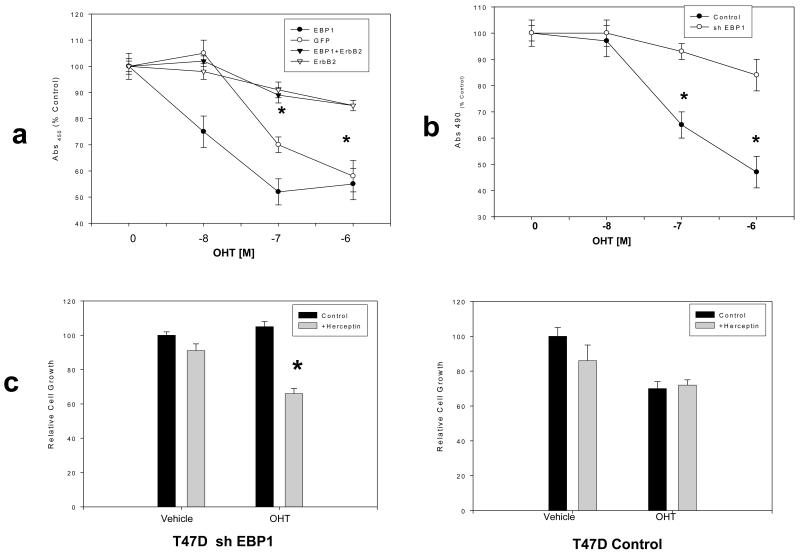
We first tested if ectopic expression of EBP1 could enhance tamoxifen sensitivity. We found that sensitivity to low doses of tamoxifen was significantly enhanced in T47D EBP1 transfectants (Fig. 6a). To determine if this enhanced sensitivity was due to a decrease in ErbB2 levels, we transiently transfected an expression plasmid for ErbB2. This resulted in a reversal of the effects of EBP1 on tamoxifen sensitivity and induction of tamoxifen resistance (Fig.6a) as previously demonstrated.
Fig. 6. Resistance of EBP1 knock out cells to tamoxifen is mediated by ErbB2.
a) T47D vector control, EBP1 transfected cells, or these cells transiently transfected with an ErbB2 expression vector were treated with the indicated concentrations of OHT. Viable cells were quantified four days later using a Promega Cell Proliferation assay. Each data point represents the Mean ±S.D. of 6 wells. Similar results were observed in two independent experiments *=differences between control and ErbB2 transfected cells at indicated concentrations of OHT were significant at P<0.05. b) T47D cells stably transduced with a control lentivirus (control) or an shRNA EBP1 targeted lentivirus (clone 4, Fig.1C), were incubated for 4 days in the presence of OHT at the indicated concentrations or vehicle control. Viable cells were quantified using a Promega Cell Proliferation assay. Each data point represents the Mean ±S.D. of 6 wells. Similar results were observed in two independent experiments. c) EBP1 ablated T47D cells (left panel) or T47D controls (right panel) were treated with OHT (10−7M) and/or Herceptin (250μg/ml) as indicated. Relative cell growth was determined by MTT assays at Day 4. 100% growth=growth in the absence of either OHT or Herceptin. In b and c*P<0.05 compared with control vector; #P<.01 compared with control vector.
We previously showed that elimination of EBP1 induced tamoxifen resistance in MCF-7 cells and upregulated ErbB2 expression [21]. Similarly, Fig. 1C shows that ErbB2 expression was upregulated after elimination of EBP1 expression in T47D cells. We therefore determined if silencing of EBP1 could also induce tamoxifen resistance in T47D cells. EBP1 silenced cells were treated with OHT and cellular sensitivity assessed. EBP1 knock out cells (now ER+ and ErbB2 high) were no longer sensitive to OHT (Fig. 6b). To determine if the tamoxifen resistance that occurred after EBP1 silencing was due primarily to the increase in ErbB2 levels, we treated EBP1 knock out T47D cells with the anti-ErbB2 antibody Herceptin in addition to OHT. We found that Herceptin restored sensitivity to tamoxifen that had been lost due to EBP1 silencing (Fig. 6c, left panel), suggesting EBP1’s primary mechanism of action may be via decreases of ErbB2 levels. In contrast, Herceptin treatment had no effect on the growth or the response to tamoxifen of T47D cells transduced with a control lentivirus (Fig.6c, right panel).
DISCUSSION
Increasing data support the clinical importance of specific ErbB heterodimers and their interacting partners in breast cancer development [2]. We have previously shown that the ErbB3 binding protein EBP1 inhibited the growth of ErbB2/ ErbB3 expressing breast cancer cell lines in response to HRG in part by downregulating protein levels of ErbB2 [21]. We now show here that the EBP1-induced inhibition of ErbB2 protein was due to changes at the level of both ErbB2 transcription and protein stability.
The ability of EBP1 to regulate ErbB2 at multiple levels was somewhat surprising. However, EBP1 has previously been demonstrated by both biological and structural analysis to be multifunctional [30]. The crystal structure of EBP1 [30] reveals that it has domains that may interact with DNA, RNA and protein. Indeed, many independent groups have demonstrated that EBP1 binds DNA [31, 32], RNA [33-36] and proteins [15, 37, 38] with varying biological consequences. What controls this differential activity of EBP1 is unknown. EBP1 is a phosphoprotein and it is possible that specific post translational modifications may result in activation of different functions. For example, we have shown that phosphorylation of EBP1 at Ser363 is essential for its ability to bind DNA through interactions with Sin3A [39]. Extensive data from Ye’s group indicate that phosphorylation of EBP1 at Ser 360 affects its interaction with nuclear AKT [37], caspase 3[40] and Bre1 [28].
We first found that steady state mRNA levels of ErbB2 were decreased in EBP1 transfected cells. Possible changes in transcriptional rate were suggested by studies indicating that EBP1 decreased the activity of a luciferase reporter construct containing the ErbB2 proximal promoter in cells that overexpress ErbB2. However, the activity of this promoter construct was not decreased by ectopic expression of EBP1 in cells with normal levels of ErbB2 expression, although mRNA levels were decreased. It is possible that a different complement of transcriptional activators exist in cells that express low levels of ErbB2 that are differentially affected by EBP1. Currently, we do not know if EBP1 inhibits the ErbB2 promoter directly, or through interactions with other proteins. EBP1 is a member of the SF 00553 protein family of DNA binding proteins, and thus may be predicted to bind directly to the ErbB2 promoter alone or in a complex of nuclear proteins. We previously showed via ChIP and EMSA analysis that EBP1 can directly bind to promoters containing E2F1 [19] or Androgen Response Elements [20, 24]. In addition, it is possible that EBP1 may interact with other transcriptional activators or repressors to decrease ErbB2 transcription. The −500 bp region of the ErbB2 promoter contains an Ap2γ site. Proteins such as Wwox have been shown to inhibit ErbB2 transcription by interfering with Ap2γ recruitment [41]. Whether or not EBP1 interacts with Wwox and Ap2γ remains to be determined.
The protein levels of ErbB2 in both BT474 and T47D EBP1 transfectants cannot be wholly accounted for by changes in mRNA levels. Thus, some additional EBP1 regulated processes may govern final protein levels. We unexpectedly found that ErbB2 protein stability was changed in EBP1 transfectants.. EBP1 has no consensus sites for ubiquitin ligase activity. In addition, EBP1 does not bind directly to human ErbB2 ectopically expressed in murine cells [15] or endogenous ErbB2 in BT474 cells (data not shown). However, it is possible that EBP1 enhances the expression, activity or recruitment of E3 ligases such as Chip [27] or LRIG [42] that are important in ErbB2 ubiquitination and degradation. Of interest, EBP1 itself is subjected to ubiquitin mediated degradation. EBP1 protein has two forms: a p42 form which is a tumor suppressor and a p48 form that is a tumor promoter [43]. E3 ubiquitin ligase hBRE1 can promote EBP1 ubiquitination and degradation in prostate cancer cell lines[28]. However, only the p42 tumor suppressor form is affected in human prostate cancer cells. In our studies, inhibition of proteasome activity did not induce the appearance of the p42 band, nor increase expression of the p48 form.
We have previously found that knock out of EBP1 in MCF-7 cells results in tamoxifen resistance. We similarly found that inhibition of EBP1 expression renders T47D cells tamoxifen resistant. To further explore the mechanism of this inhibiton, we found that blocking ErbB2 activity with Herceptin reversed the EBP1 induced tamoxifen resistance. Thus, we postulate that EBP1 may regulate tamoxifen sensitivity in part by modulating levels of ErbB2. Similarly, the transcription factor PAX2 represses ErbB2 transcription by competing with SRC-3 for binding to Estrogen response elements within an intronic region of the ErbB2 gene. The repression of ErbB2 transcription by PAX 2 results in tamoxifen sensitivity [44]..
In summary, we have found that ectopic expression of EBP1 suppresses ErbB2 protein levels in ER+ human breast cancer cell lines that express ErbB2 at both high and low levels. EBP1 affected both transcription of ErbB2 mRNA and ErbB2 protein levels by altering ErbB2 stability and enhancing ubiquitination. Abrogation of EBP1 protein expression rendered T47D cells tamoxifen resistant and this resistance was mediated via the ErbB2 pathway. Thus, restoration of EBP1 or EBP1 signaling pathways may provide a new alternative in hormone refractory breast cancer.
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01 CA76047, RC1 CA145066-01 and DOD W81XWH-08-1-0560 (to AWH) and by the Project of the Shanghai Science and Technology Committee (08JC1414400, 08JC1404800) and the Shanghai Leading Academic Discipline Project (S30206). We thank Dr. Kay Huebner, Ohio State University Cancer Center,Dr. Helen Hurst for the ErbB2 promoter plasmid and Dr. Yossi Yarden for the ErbB2 expression construct.
References
- 1.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 2.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 4.Lemoine NR, Barnes DM, Hollywood DP, Hughes CM, Smith P, Dublin E, Prigent SA, Gullick WJ, Hurst HC. Expression of the ERBB3 gene product in breast cancer. Br J Cancer. 1992;66:1116–1121. doi: 10.1038/bjc.1992.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiseman SM, Makretsov N, Nielsen TO, Gilks B, Yorida E, Cheang M, Turbin D, Gelmon K, Huntsman DG. Coexpression of the type 1 growth factor receptor family members HER-1, HER-2, and HER-3 has a synergistic negative prognostic effect on breast carcinoma survival. Cancer. 2005;103:1770–1777. doi: 10.1002/cncr.20970. [DOI] [PubMed] [Google Scholar]
- 6.Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, Moasser MM. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445:437–441. doi: 10.1038/nature05474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin BJ, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 8.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, III, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedard PL, Freedman OC, Howell A, Clemons M. Overcoming endocrine resistance in breast cancer: are signal transduction inhibitors the answer? Breast Cancer Res Treat. 2008;108:307–317. doi: 10.1007/s10549-007-9606-8. [DOI] [PubMed] [Google Scholar]
- 10.Piccart-Gebhart MJ, Procter M, Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga J, Bell R, Jackisch C, Cameron D, Dowsett M, Barrios CH, Steger G, Huang CS, Andersson M, Inbar M, Lichinitser M, Lang I, Nitz U, Iwata H, Thomssen C, Lohrisch C, Suter TM, Ruschoff J, Suto T, Greatorex V, Ward C, Straehle C, McFadden E, Dolci MS, Gelber RD. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med. 2005;353:1659–1672. doi: 10.1056/NEJMoa052306. [DOI] [PubMed] [Google Scholar]
- 11.Pietras RJ, Arboleda J, Reese DM, Wongvipat N, Pegram MD, Ramos L, Gorman CM, Parker MG, Sliwkowski MX, Slamon DJ. HER-2 tyrosine kinase pathway targets estrogen receptor and promotes hormone-independent growth in human breast cancer cells. Oncogene. 1995;10:2435–2446. [PubMed] [Google Scholar]
- 12.Benz CC, Scott GK, Sarup JC, Johnson RM, Tripathy D, Coronado E, Shepard HM, Osborne CK. Estrogen-dependent, tamoxifen-resistant tumorigenic growth of MCF-7 cells transfected with HER2/neu. Breast Cancer Res Treat. 1992;24:85–95. doi: 10.1007/BF01961241. [DOI] [PubMed] [Google Scholar]
- 13.Kraus MH, Issing W, Miki T, Popescu NC, Aaronson SA. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci U S A. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plowman GD, Whitney GS, Neubauer MG, Green JM, McDonald VL, Todaro GJ, Shoyab M. Molecular cloning and expression of an additional epidermal growth factor receptor-related gene. Proc Natl Acad Sci U S A. 1990;87:4905–4909. doi: 10.1073/pnas.87.13.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo JY, Wang XW, Rishi AK, Lessor T, Xia XM, Gustafson TA, Hamburger AW. Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br J Cancer. 2000;82:683–690. doi: 10.1054/bjoc.1999.0981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lessor TJ, Yoo JY, Xia X, Woodford N, Hamburger AW. Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J Cell Physiol. 2000;183:321–329. doi: 10.1002/(SICI)1097-4652(200006)183:3<321::AID-JCP4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 17.Xia X, Cheng A, Lessor T, Zhang Y, Hamburger AW. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J Cell Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- 18.Zhang YX, Woodford N, Xia XM, Hamburger AW. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Research. 2003;31:2168–2177. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Hamburger AW. Heregulin regulates the ability of the ErbB3-binding protein Ebp1 to bind E2F promoter elements and repress E2F-mediated transcription. J Biol Chem. 2004;279:26126–26133. doi: 10.1074/jbc.M314305200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, Akinmade D, Hamburger AW. The ErbB3 binding protein Ebp1 interacts with Sin3A to repress E2F1 and AR-mediated transcription. Nucleic Acids Res. 2005;33:6024–6033. doi: 10.1093/nar/gki903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Akinmade D, Hamburger AW. Inhibition of heregulin mediated MCF-7 breast cancer cell growth by the ErbB3 binding protein EBP1. Cancer Lett. 2008;265:298–306. doi: 10.1016/j.canlet.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Linn D, Liu Z, Melamed J, Tavora F, Young CY, Burger AM, Hamburger AW. EBP1, an ErbB3-binding protein, is decreased in prostate cancer and implicated in hormone resistance. Mol Cancer Ther. 2008;7:3176–3186. doi: 10.1158/1535-7163.MCT-08-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JS, Gong A, Cheville JC, Smith DI, Young CY. AGR2, an androgen-inducible secretory protein overexpressed in prostate cancer. Genes Chromosomes Cancer. 2005;43:249–259. doi: 10.1002/gcc.20188. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, Goloubeva O, Ross DD, Brodie A, Hamburger AW. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc Natl Acad Sci U S A. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lu Y, Zhou H, Lee M, Liu Z, Hassel BA, Hamburger AW. Alterations in cell growth and signaling in ErbB3 binding protein-1 (Ebp1) deficient mice. BMC Cell Biol. 2008;9:69. doi: 10.1186/1471-2121-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Oh SM, Okada M, Liu X, Cheng D, Peng J, Brat DJ, Sun SY, Zhou W, Gu W, Ye K. Human BRE1 is an E3 ubiquitin ligase for Ebp1 tumor suppressor. Mol Biol Cell. 2009;20:757–768. doi: 10.1091/mbc.E08-09-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klapper LN, Waterman H, Sela M, Yarden Y. Tumor-inhibitory antibodies to HER-2/ErbB-2 may act by recruiting c-Cbl and enhancing ubiquitination of HER-2. Cancer Res. 2000;60:3384–3388. [PubMed] [Google Scholar]
- 30.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada H, Mori H, Momoi H, Nakagawa Y, Ueguchi C, Mizuno T. A fission yeast gene encoding a protein that preferentially associates with curved DNA. Yeast. 1994;10:883–894. doi: 10.1002/yea.320100704. [DOI] [PubMed] [Google Scholar]
- 32.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38-2G4, which varies with the cell cycle. Exp Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 33.Squatrito M, Mancino M, Sala L, Draetta GF. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem Biophys Res Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 34.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 35.Bose SK, Sengupta TK, Bandyopadhyay S, Spicer EK. Identification of Ebp1 as a component of cytoplasmic bcl-2 mRNP (messenger ribonucleoprotein particle) complexes. Biochem J. 2006;396:99–107. doi: 10.1042/BJ20051548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 37.Ahn JY, Liu X, Liu Z, Pereira L, Cheng D, Peng J, Wade PA, Hamburger AW, Ye K. Nuclear Akt associates with PKC-phosphorylated Ebp1, preventing DNA fragmentation by inhibition of caspase-activated DNase. EMBO J. 2006;25:2083–2095. doi: 10.1038/sj.emboj.7601111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okada M, Jang SW, Ye K. Ebp1 association with nucleophosmin/B23 is essential for regulating cell proliferation and suppressing apoptosis. J Biol Chem. 2007;282:36744–36754. doi: 10.1074/jbc.M706169200. [DOI] [PubMed] [Google Scholar]
- 39.Akinmade D, Lee M, Zhang Y, Hamburger AW. Ebp1-mediated inhibition of cell growth requires serine 363 phosphorylation. Int J Oncol. 2007;31:851–858. [PubMed] [Google Scholar]
- 40.Liu Z, Liu X, Nakayama KI, Nakayama K, Ye K. Protein kinase C-delta phosphorylates Ebp1 and prevents its proteolytic degradation, enhancing cell survival. J Neurochem. 2007;100:1278–1288. doi: 10.1111/j.1471-4159.2006.04313.x. [DOI] [PubMed] [Google Scholar]
- 41.Qin HR, Iliopoulos D, Nakamura T, Costinean S, Volinia S, Druck T, Sun J, Okumura H, Huebner K. Wwox suppresses prostate cancer cell growth through modulation of ErbB2-mediated androgen receptor signaling. Mol Cancer Res. 2007;5:957–965. doi: 10.1158/1541-7786.MCR-07-0211. [DOI] [PubMed] [Google Scholar]
- 42.Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, III, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 43.Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc Natl Acad Sci U S A. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS. Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature. 2008;456:663–666. doi: 10.1038/nature07483. [DOI] [PMC free article] [PubMed] [Google Scholar]