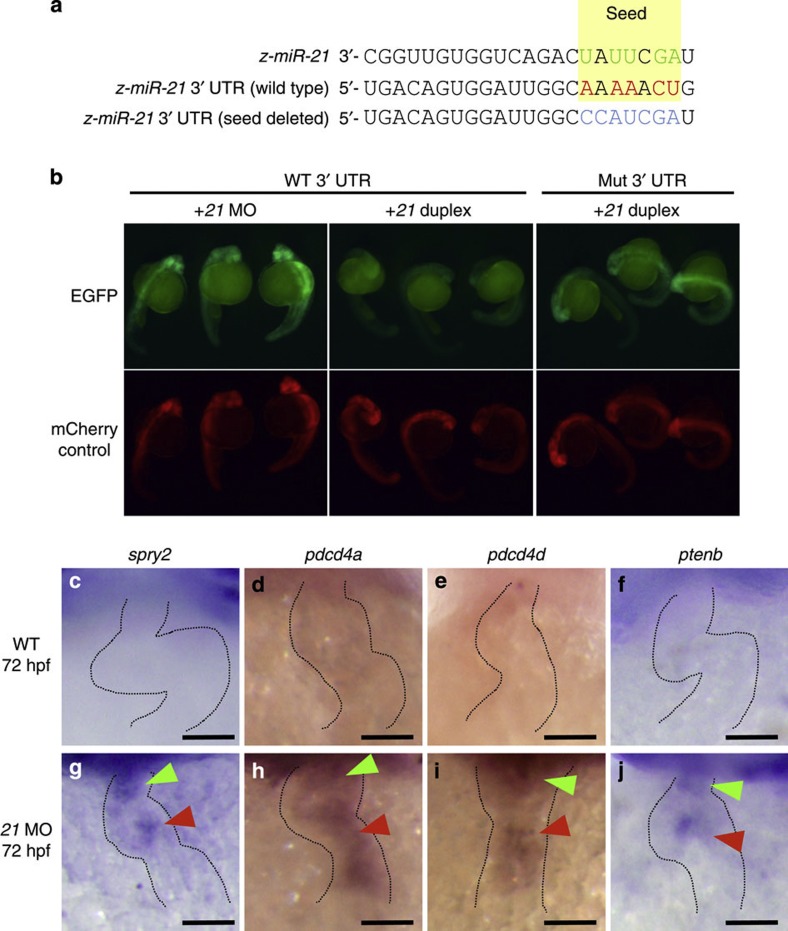
Figure 5. Sequence alignment and expression patterns of selected genes.
(a) Sequence alignment of zebrafish miR-21, 3′-UTR of spry2 and its mutated version is shown, with the seed sequences highlighted in yellow and the complementary nucleotides in red and green. (b) The EGFP-spry2 reporter with the WT spry2 3′-UTR gave strong fluorescent signals when the endogenous miR-21 was blocked by the MO against it (left panel), but failed to show the signals when co-injected with the miR-21 duplex (middle panel). The EGFP-spry2 reporter mRNA, in which the seed sequence was mutated, produced strong signals even when co-injected with the miR-21 duplex (right panel) at 24 hpf. (c,g) Expression of the endogenous spry2 was not observed in the WT at 72 hpf (c), whereas it was induced in both the OFT and AV canal in the morphant (green and red arrowheads in g, respectively). (d–j) Other target genes, pdcd4a (d,h), pdcd4b (e,i) and ptenb (f,j) were induced in the morphant (h–j). Expression of these genes was not observed in the WT (c–f). Scale bars, 50 μm (c–j).