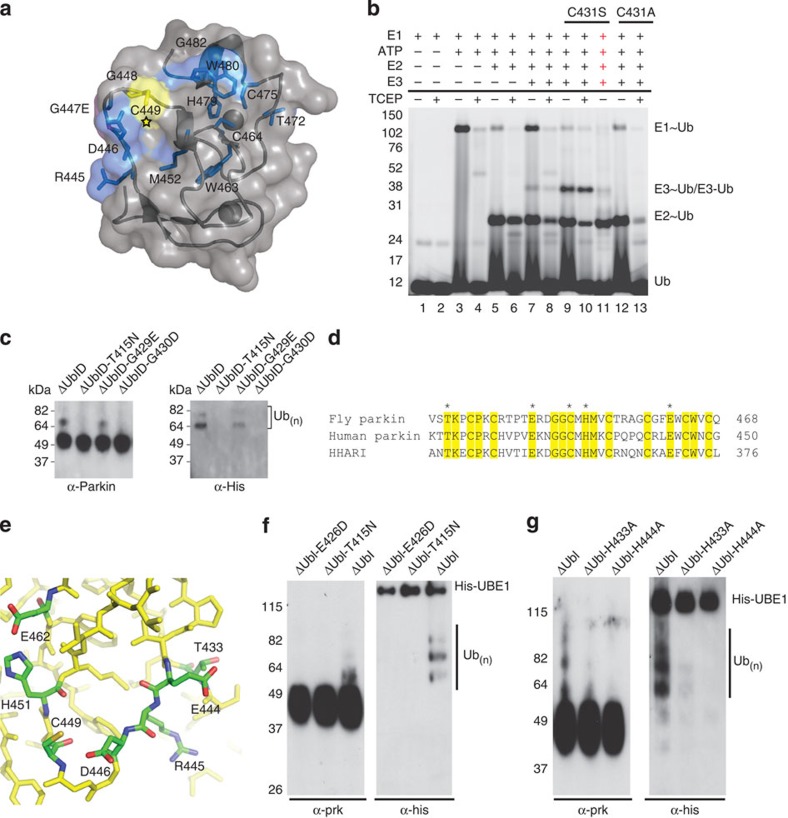
Figure 3. Pathogenic mutations in parkin RING2.
The structure of parkin predicts catalytic residues conserved between parkin and HHARI. (a) The structure of parkin with the catalytic cysteine highlighted (star) and chemical-shift perturbation experiments show affected residues (blue) in parkin RING2 by the ARJP substitution G447E (magenta). (b) The formation of a reducible parkin~Ub thiolester was monitored using His-SUMO-IBR-R2-parkin. Lanes 1–2 contain E1, Mg2+ and Cy5–ubiquitin incubated for 10 min at 37 oC. Lanes 3–4 have ATP added for 10 min. Lanes 5–6 have E2 added followed by a further 10-min incubation. Lanes 7–8 have wild-type parkin added. Lanes 9–11 have C431S–parkin added, with NaOH added to lane 11, indicated by red crosses. Lanes 12–13 have C431A–parkin added; ‘−’ and ‘+’ below the line indicate the absence or presence of TCEP. (c) An autoubiquitination assay of folded pathogenic mutants. Parkin–ubiquitin conjugates are detected by western blotting using parkin (left) and His-ubiquitin (right) antibodies. (d) An alignment of the RING2 domains of fly and human parkin, and HHARI. Conserved residues are shaded yellow and the potential catalytic residues are marked with an asterisk. (e) Close-up view of the surface of parkin RING2 with the potential catalytic residues indicated. (f) Autoubiquitination assay of active parkin (ΔUbl) and parkin ΔUbl-E426D. (g) Autoubiquitination assays of active parkin (ΔUbl) and parkin ΔUbl-H433A and ΔUbl-H444A. In f and g, parkin–ubiquitin conjugates are detected by western blotting using parkin (left) and His-ubiquitin (right) antibodies.