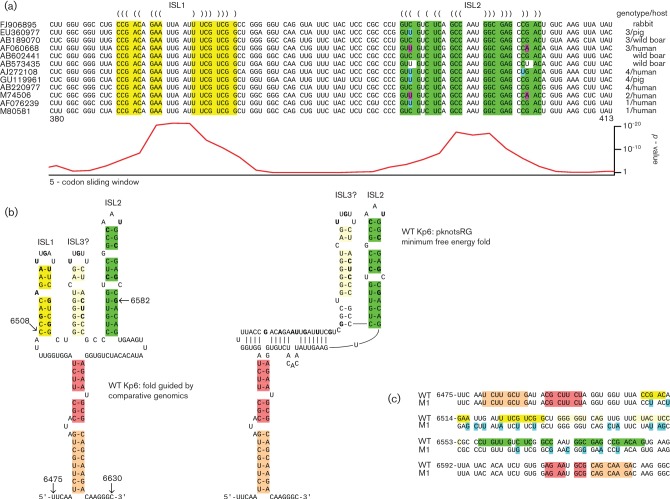
Fig. 2.
Identification of potential RNA secondary structure in the central conserved region in ORF2. (a) Extracts from representative sequences showing the predicted ISL1 (yellow) and ISL2 (green) stem–loop structures in the conserved region. Predicted base pairings are indicated with parentheses. Single substitutions that preserve the predicted base pairings (e.g. G-C to G-U) are indicated with cyan. Paired substitutions that preserve the predicted base pairings are indicated with pink. GenBank accession numbers are shown at left. Genotype and isolation source species are shown at right. See Johne et al. (2012) for a phylogenetic tree. Numbers 380 and 413 below indicate codon positions in ORF2. Note that, in some isolates, ISL1 and ISL2 can be basally extended by, respectively, another one or two base pairings. The corresponding synonymous site conservation P-values from Fig. 1 are shown at bottom. See also Fig. S1 for the ISL1, ISL2 and flanking sequences from all 205 ORF2 sequences used. (b) Predicted secondary structure of the ISL1+ISL2 and flanking region from isolate Kp6. Structures ISL1 and ISL2 (left) are based on comparative genomics. In Kp6 and some other isolates, however, the minimum free energy fold (right) contains ISL2 and the peripheral base pairings (indicated with orange/salmon), but ISL1 is disrupted and replaced by a new stem–loop, ISL3. Nucleotides that were altered in the M1 mutant are indicated in bold. Nucleotide coordinates refer to GenBank accession JQ679013. Nucleotides 6508 to 6582 correspond to the region inserted into the HVR. (c) In the M1 mutant, many synonymous mutations (cyan) were introduced within the two regions showing highest conservation (corresponding to ISL1 and ISL2), and additional mutations were introduced to also disrupt the potential ISL3. The colour coding in the WT sequence corresponds to the structure predictions in Fig. 2b. Numbers at left indicate genomic coordinates of the first nucleotide in each line.