Abstract
In order to successfully cure patients with prostate cancer (PCa), it is important to detect the disease at an early stage. The existing clinical biomarkers for PCa are not ideal, since they cannot specifically differentiate between those patients who should be treated immediately and those who should avoid over-treatment. Current screening techniques lack specificity, and a decisive diagnosis of PCa is based on prostate biopsy. Although PCa screening is widely utilized nowadays, two thirds of the biopsies performed are still unnecessary. Thus the discovery of non-invasive PCa biomarkers remains urgent. In recent years, the utilization of urine has emerged as an attractive option for the non-invasive detection of PCa. Moreover, a great improvement in high-throughput “omic” techniques has presented considerable opportunities for the identification of new biomarkers. Herein, we will review the most significant urine biomarkers described in recent years, as well as some future prospects in that field.
Keywords: prostate cancer, biomarker, urine, non-invasive
1. Introduction
Cancer is one of the most critical health problems in our society, both in terms of morbidity and social impact. Prostate cancer (PCa) is the most commonly diagnosed cancer among European and American men (29% of all cases) [1,2]. Although PCa is a slow growing tumor that affects older men, it is still a lethal disease and is currently the second most common cause of cancer death among men [2]. The long latency period of this type of cancer and its potential curability make this disease a perfect candidate for screening [3].
Current screening techniques are based on a measurement of serum prostate specific antigen (PSA) levels and a digital rectal examination (DRE). A decisive diagnosis of PCa is based on transrectal ultrasound-guided prostate biopsies (PBs). The use of serum PSA as a cancer-specific detection test has some well-recognized limitations, such as a low positive predictive value (PPV).
When PSA is 4.0–10.0 ng/mL, the PPV is 18% to 25% (mean, 21%), and when PSA is >10 ng/mL, the PPV is 58% to 64% (mean, 61%), when combined with a DRE as a screening tool this still results in approximately 66% negative PBs [4–6]. These patients are often subjected to repeat PSA measurements and PBs (the “over-diagnosis” problem). “Over-treatment,” through the detection of non-life-threatening tumors [7], especially in the so-called gray zone (serum PSA between 4–10 ng/mL), represents yet another dilemma, as it is difficult to discriminate between patients with PCa and those with benign prostatic hyperplasia (BPH) or between those patients suffering from prostatitis and the results of urethral manipulation, which can also increase PSA levels [8]. Conversely, the prevalence of PCa in patients with PSA levels below the threshold of 4 ng/mL is around 15% resulting in undiagnosed cases of the disease [9,10]. As a consequence of the current screening parameters, approximately two thirds of the 1 million biopsies made annually both in the United States and in Europe are unnecessary [1,2]. There is therefore an urgent need for new and more effective biomarkers for PCa that can help to better identify which patients should undergo further diagnostic tests and also help to detect which patients will develop an aggressive tumor and, therefore, will need immediate treatment.
2. Urine: A Source of Prostate Cancer Biomarkers
The discovery of biomarkers is based on the following research principle: the comparison of physiological states, phenotypes or changes across control and case (disease) patient groups [11]. A key approach to biomarker discovery is to compare case versus control samples in order to detect statistical differences that can lead to the identification and prioritization of potential biomarkers. Theoretically, this could be a single biomarker molecule, however, it is more likely to be a panel of up- and down- regulated molecules and/or proteins with altered post-translational modifications (PTMs) that differ in normal and disease states [12,13]. Here we have focused our biomarker classification system on the basis of their potential applications for screening, diagnosis, prognosis or prediction (see Box I).
Box I. Types of Biomarkers Based on their Applications.
Screening/detection biomarkers, like serum PSA, are used to predict the potential occurrence of disease in asymptomatic men or those with non-disease-specific symptoms.
Diagnostic biomarkers are used to make predictions for patients suspected of having a disease. An ideal diagnostic biomarker should enable an unbiased conclusion, particularly in patients without specific symptoms. It should fulfill several criteria: (i) high specificity for a given disease (low rate of false positives); (ii) high sensitivity (low rate of false negatives); (iii) ease of use (rapid procedure); (iv) standardization (consistent reproducibility); (v) clearly readable result for clinicians [13]; (vi) cost-effectiveness; and (vii) ability to be quantified in an accessible biological fluid or sample.
Prognostic biomarkers are used to predict the overall outcome of a patient, regardless of therapy.
Predictive biomarkers are used to identify subpopulations of patients who are most likely to respond to a given therapy. A predictive biomarker can be a target for therapy.
In recent years interest in searching for new biomarkers obtainable by non-invasive means has increased significantly. For centuries, physicians have attempted to use urine for the non-invasive assessment of disease. Urine is produced by the kidneys and allows the human body to eliminate waste products from the blood. Urine may contain information not only from kidney and urinary tracts, but also from distant organs via plasma obtained through glomerular filtration. The analysis of urine could, therefore, allow the identification of biomarkers for both urogenital and systemic diseases.
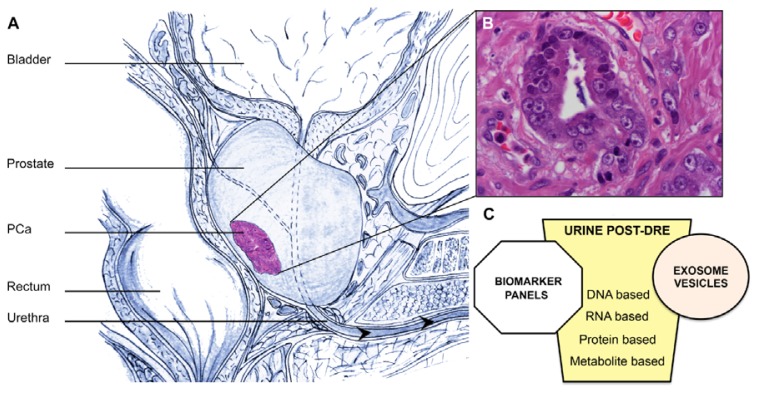
The main function of the prostate gland is the secretion of prostatic fluid, which on ejaculation is combined with seminal vesicle derived fluid to promote sperm activation and function [14]. The gentle massage of each side of the prostate gland during DRE stimulates the release and movement of prostatic fluids and detached epithelial cells into the urethra [14] (Figure 1). These fluids can contain both cells and secretions originating in PCa [15]. PCa cells were first described in voided urine by Papanicolaouin 1958 [16], however they appear to be fragile and low in number [17] underlying the need for careful collection, manipulation and storage of urine prior to analysis. Urine collection can be accomplished without a disruption of standard clinical practice and can be sampled multiple times throughout the course of prostatic disease. Nevertheless, using urine for the discovery of biomarkers presents some important technical challenges.
Figure 1.
(A) Anatomical location of the prostate; (B) Prostate cancer cells; (C) Biomarkers found in urine. Based on their descriptions, biomarkers can be divided into the following groups: DNA-based, RNA-based, and protein-based. Of late, urinary exosomes, which are secreted vesicles that contain proteins and functional RNA and miRNA molecules, have emerged as a novel approach to acquiring new PCa biomarkers.
The search for effective biomarkers has principally included transcriptional profiling, DNA methylation, metabolomics, fluxonomics, and more recently, proteomics [18]. Emerging biomarkers have the potential to be developed into new and clinically reliable indicators, which will have a high specificity for the diagnosis and prognosis of PCa. Ideally biomarker acquisition will be less invasive than current clinical means, and will be useful for screening men for PCa, and be able to guide patient management to provide maximum benefits while minimizing treatment-related side effects and risks [19]. This review focuses on published data referring promising DNA, RNA, miRNA, protein and metabolite based urine biomarkers (Table 1) and highlights exosomes as a new source of PCa urinary biomarkers.
Table 1.
Summary of PCa biomarkers in the literature.
| Gene | Description | Gene type | Expression | Type of biomarker | Sample | References |
|---|---|---|---|---|---|---|
| AMACR (P504) | Alpha-Methylacyl-CoA Racemase | Enzyme involved in branched chain fatty acid oxidation | Over-expressed in PCa (also in HGPIN) and in some other carcinomas, both at RNA and protein level | Diagnostic (in gray zone) and prognostic | Tissue, blood and urine | [20,21] |
| ANXA3 | Annexin A3 | Calcium and phospholipid binding protein | Presence in urinary exosomes and proteasomes. Lower production in PCa than in BPH, HGPIN and benign | Prognostic (able to stratify a large group of intermediate-risk patients into high- and low-risk subgroups) | Tissue and urine | [22–24] |
| APC | Adenomatous polyposis coli | Tumor suppressor. Promotes rapid degradation of CTNNB1 and participates in Wnt signaling as a negative regulator. | APC methylation higher in PCa than in BPH. Methylation level correlates positively with Gleason score | Diagnostic and prognostic | Tissue and Urine DNA | [25] |
| AR | Androgen receptor | Receptor for androgen stimulation of prostate. | Over-expression associated with poor prognosis prostate cancer and metastasis | Prognostic | Tissue RNA and IHC | [26–28] |
| AURKA | Aurora kinase. | Aurora kinase. AURKA is a centrosome-associated serine/threonine kinase involved in mitotic chromosomal segregation. | Amplified and over-expressed in certain types of poor prognosis prostate cancer | Prognostic | Tissue RNA and DNA | [29–31] |
| AZGP1 | Alpha-2-glycoprotein 1, zinc binding. Alias. ZAG | Stimulates lipid degradation in adipocytes and causes the extensive fat losses associated with some advanced cancers. May bind polyunsaturated fatty acids. | Over-expressed in PCa. Low AZGP1 expression predicts for recurrence in margin-positive, localized PCa | Diagnostic, prognostic | Tissue, blood and urine | [32–34] |
| BRAF | v-raf murine sarcoma viral oncogene homolog B1 | Belongs to the raf/mil family of serine/threonine protein kinases and is involved in the regulation of the MAP kinase/ERKs signaling pathway, which affects cell division, differentiation. | SLC45A3-BRAF fusion gene, mutations and gain in prostate cancer | Diagnostic and therapeutic target | Tissue RNA and DNA | [35–37] |
| CAMKK2 | Calcium/calmodulindependent protein kinase kinase 2. | AR target gene promoting biosynthesis and glycolysis | Down-regulation of calcium/calmodulin-dependent protein kinase kinase 2 by androgen deprivation induces castration-resistant prostate cancer. | Prognostic | Tissue RNA | [38–40] |
| CDH1 | Cadherin 1, type 1, E-cadherin (epithelial) | Epithelial cell - cell adhesion molecule | Reduced production in 50% of tumors. E-cadherin production by epithelial cells has been shown to predict PCa prognosis | Prognostic (correlated with grade, tumor stage, and survival) | Tissue | [41,42] |
| CLU | Clusterin | Function unknown, but is thought to be involved in several basic biological events such as cell death and tumor progression. | Developed as a potential therapeutic target | Therapeutic target | Tissue, exosome protein | [43–46] |
| CRISP-3 | Cysteine-Rich Secretory Protein 3 | Secreted protein produced in the male reproductive tract, is involved in sperm maturation | Large amounts have been detected in seminal plasma. Over-expressed in HGPIN and PCa. | Prognostic | Tissue | [47,48] |
| EPCA | Early Prostate Cancer Antigen | Nuclear matrix protein | Over-expressed in PCa | Diagnostic (for predicting repeated BP) | Tissue and blood | [49,50] |
| EPCA-2 | Early Prostate Cancer Antigen 2 | Nuclear matrix protein | Over-expressed in PCa | Diagnostic and Prognostic (differentiate localized PCa from metastatic PCa) | Blood | [51] |
| FOLH1/PS MA | Folate hydrolase 1/Prostate Specific Membrane Antigen | Type II membrane protein. 1/N-acetylated-alpha-linked acidic dipeptidase | Over-expressed in PCa compared to BPH and normal | Diagnostic. Imaging marker and target for therapy | Tissue, blood and urine | [52,53] |
| GOLM1 | Golgi membrane protein 1 (GOLPH2) | Cis-Golgi membrane protein of unknown function | Over-expressed in PCa | Diagnostic | Urine | [54,55] |
| GSTP1 | Glutathione S-transferase P1 | Enzyme involved in protecting DNA from free radicals | Loss of GSTP1 expression due to the promoter hypermethylation (>90% of PCa). Correlates with the number of cores found to contain PCa | Diagnostic (indicator for repeat biopsy) | Tissue and urine DNA | [56,57] |
| HPN | Hepsin | Membrane serine protease | Over-expressed in 90% PCa tumors (highly produced in HGPIN and PCa compared with BPH) | Diagnostic | Tissue | [58,59] |
| IL-6 | Interleukin-6 | Cytokine secreted by a variety of cell types, is involved in the immune and acute-phase response | Increased concentrations of IL-6 and IL-6R in metastatic and androgen-independent PCa | Diagnosis and Prognostic | Blood | [60–62] |
| IMPDH2 | IMP (inosine 5′-monophosphate) dehydrogenase 2 | Myc target gene associated with nucleotide biosynthesis | Increased serum level associated with the clinicopathological features of the patients with PCa | Diagnostic | Blood | [63] |
| KLK2 | Human Kallikrein 2 | Secreted serine protease | Over-expressed during PCa progression | Diagnostic and Prognostic | Tissue and blood | [64,65] |
| KLK3 (PSA) | Kallikrein-related peptidase 3 (Prostate-Specific Antigen) | Secreted serine protease. Serum level of this protein, called PSA in the clinical setting, is useful in the diagnosis and monitoring of PCa. | Increased expression associated with malignant PCa | Diagnostic | Blood, urine | [66] |
| KLK4 | Kallikrein-related peptidase 4 | One of fifteen kallikrein subfamily members located in a cluster on chromosome 19 | Increased expression associated with malignant PCa | Prognostic | Tissue RNA and IHC | [67,68]. |
| MAP3K5 | Mitogen-activated protein kinase kinase kinase 5 | Signaling cascade | Increased expression associated with PCa | Prognostic | Tissue RNA and IHC | [69] |
| MKI67 | Encoding antigen identified by monoclonal antibody Ki-67 | Tumor growth marker, encodes a nuclear protein that is associated with and may be necessary for cellular proliferation | Increased expression associated with malignant prostate cancer | Prognostic | Tissue | [70–72] |
| MMP26 | Matrix metallo peptidase 26 | Involved in the breakdown of extracellular matrix in normal physiological processes and cancer metastasis. | Highest expression in HGPIN and decline in cancer, possible involvement in formation of early cancer. | Progression | Tissue RNA | [73–76] |
| MMP9 | Matrix metallo proteinase 9 | Implicated in invasion and metastasis of human malignancies | Over-expressed in PCa | Diagnostic | Urine | [77,78] |
| OR51E2/PS GR | Prostate Specific G-coupled Receptor | Receptors coupled to heterotrimeric GTP-binding proteins | Over-expressed in PCa | Diagnostic | Tissue and urine | [79–81] |
| PAP | Human Prostatic acid phosphatase | Enzyme | Over-expressed in PCa and in bone metastasis | Diagnostic and Prognostic of PCa bone metastasis | Blood and urine | [82,83] |
| PCA3 | Prostate Cancer Gene 3 | Non coding mRNA | Prostate specific and highly up-regulated in PCa | Diagnostic (indicator for repeat biopsy) | Tissue and urine | [84–93] |
| PDIA3 | Protein disulfide isomerase family A, member 3. | Endoplasmic reticulum that interacts with lectin chaperones calreticulin and calnexin to modulate folding of newly synthesized glycoproteins. | Increased expression associated with malignant PCa | Prognostic | Tissue RNA and IHC | [69] |
| PSCA | Prostate Stem Cell Antigen | Membrane glycoprotein | Specific production in the prostate and possible target for therapy | Prognostic (correlated with higher Gleason score, higher stage, and the presence of metastasis) | Tissue and blood | [94,95] |
| RARB | Retinoic acid receptor, beta | Binds retinoic acid. Mediates signalling in embryonic morphogenesis, cell growth and differentiation. | DNA methylation | Prognostic | Tissue and urine DNA | [96,97] |
| RASSF1A | Ras association (RalGDS/AF-6) domain family member 1 | Potential tumor suppressor. Required for death receptor-dependent apoptosis | DNA methylation | Prognostic | Tissue and urine DNA | [97] |
| Sarcosine | Sarcosine | N-methyl derivative of the amino-acid glycine | Seems to be differentially expressed metabolite elevated during PCa progression to metastasis | Prognostic | Urine and blood | [98] |
| SPINK1 | Serine peptidase inhibitor, Kazal type 1 | Serine peptidase inhibitor | Overexressed in a portion of non-ETS translocated tumors | Diagnostic | Urine, tissue | [54,99] |
| TERT | Telomerase reverse transcriptase | Maintains the telomeric ends of chromosomes and if telomerase is active, cancer cells may escape cell cycle arrest and replicative senscence | Amplification in PCa, significative association with Gleason score | Prognostic | Urine and blood | [57,100,101] |
| TGFB1 | Transforming growth factor-b1 | Growth factor involved in cellular differentiation, immune response, angiogenesis, and proliferation | Role of TGFβ1 in PCa progression. | Prognostic (Correlation with tumor grade and stage and lymph node metastasis) | Tissue and blood | [62,102,103] |
| TIMP4 | TIMP metallopeptidase inhibitor 4 | Inhibitors of the matrix metallo proteinases | Highest expression in HGPIN and decline in cancer, possible involvement in formation of early cancer. | Progression | Tissue RNA | [73–75] |
| TMPRSS2:E RG | 5′ UTR of the prostate-specific androgen regulated transmembrane protease serine2 and v-ETS erythroblostosis virus E26 oncogene homolog | Gene fusion; androgen drives the expression of ETS-TF and causes tumor proliferation | The most common gene fusion in PCa. Over-expressed PCa and related to PCa aggressiveness | Prognostic for aggressive PCa and detection of PCa | Tissue and urine | [104–106] |
| PLAU and UPAR | Plasminogen Activator, Urokinase and Receptor | Degradation of extra cellular matrix | Over-expressed in BPH and PCa vs benign | Prognostic (increased uPA and uPAR in PCa patients with bone metastasis) | Tissue and blood | [107,108] |
2.1. DNA-Based Urinary Biomarkers
DNA-based biomarkers include single nucleotide polymorphisms (SNPs), chromosomal aberrations, changes in DNA copy number, microsatellite instability, and altered promoter-region methylation [109]. The epigenetic silencing of the glutathione-S-transferase P1 (GSTP1) gene is the most common (>90%) genetic alteration so far reported in PCa [110–112]. Methylation-specific polymerase chain reaction (MSP) methods allowed the successful detection of GSTP1 methylation in urine, and ejaculates from PCa patients. A possible drawback is the high frequency of GSTP1 methylation in patients with high-grade prostatic intraepithelial neoplasia (HG PIN) and in patients with negative or suspicious PB. Further follow-up is needed to determine whether such cases are false positives or part of the significant number of under-diagnosed cancer cases in PB. Recently, Costa et al. observed significantly different methylation levels of the genes protocadherine 17 (PCDH17) and transcription factor 21 (TCF21) in PCa tissue compared to cancer free individuals, providing 83% sensitivity and 100% specificity for cancer detection. However while absolute specificity was retained in urine samples, sensitivity was only 26% [113]. In comparison, Daniunaite et al., (2011) report the high sensitivity of DNA methylation biomarkers in urine, especially that of RASSF1 (Ras association (RalGDS/AF-6) domain family member 1) and RARB (retinoic acid receptor beta) for the early and non-invasive detection of PCa. Thus, results this far suggest that methylated genes can serve as useful markers for PCa [97].
2.2. RNA-Based Urine Biomarkers
RNA-based biomarkers include coding and non-coding transcripts and regulatory RNAs, such as microRNAs (miRNAs) [109]. Improvements in RNA microarray platforms, quantitative PCR (qPCR), and the development of new high-throughput technologies, such as next-generation sequencing (NGS), allow us to better understand the expression profiles of single cells, populations of cells and specific tissues, while also allowing comparisons between different pathological conditions. In recent years, a wide range of promising PCa biomarkers that are not only prostate-specific, but also differentially expressed in prostate tumors, have been identified.
After PSA, Prostate Cancer Antigen 3 (PCA3), is the only biomarker approved by the Food and Drug Administration (FDA), and is utilised in a commercially available test under the name PROGENSA® PCA3 (Gen-Probe, San Diego, CA, USA) [84]. PCA3 was first identified in 1999 [85]. The PCA3 gene encodes a non-coding RNA (ncRNA) (see Box II) that is over-expressed in 95% of all primary PCa specimens. Some of its potential applications include testing as an alternative to a first PB and, aiding the decision whether to repeat a PB in men with high serum PSA levels and previously negative biopsies [86,87]. The measurement of PCA3 mRNA vs. PSA mRNA in urine was first proposed by Hessels et al. [88]. Later on, this study was verified in a large, European multicenter study, which concluded that PCA3 possessed potential as an aid in PCa diagnosis [89]. The assay consists of a transcription-mediated amplification, which demonstrates 69% sensitivity, 79% specificity, and an area under the curve (AUC) value of 0.75 [90]. Currently, a PCA3 score (PCA3-to-PSA ratio) cut-off of 35 has been adopted, which combines the greatest cancer sensitivity and specificity (54% and 74%, respectively) [91]. However, more recent studies have shown that a lower cut-off score of 25 might be preferable [92].
Box II. Non-coding RNA.
A “central dogma” of molecular biology was that genetic information flowed in one direction with proteins as the end product. However, growing evidence has emerged to describe the role of RNAs that are not translated into proteins. These ncRNAs comprise microRNAs, anti-sense transcripts and other transcriptional units containing a high density of stop codons and lacking any extensive “Open Reading Frame” (ORF) [139]. Several types of ncRNAs have been implicated in gene regulation via modification of the chromatin structure, alterations to DNA methylation, RNA silencing, RNA editing, transcriptional gene silencing, post-transcriptional gene silencing, and enhancement of gene expression [140–142]. It is becoming clear that these RNAs perform critical functions during development and cell differentiation [139]. The roles that small-ncRNAs, such as miRNAs and small interfering RNAs (siRNAs), play in gene silencing have been well-studied, and they have been reported to be aberrantly expressed in many cancers [140]. ncRNAs are thus emerging as a new class of functional transcripts in eukaryotes.
Prostate Specific Membrane Antigen (PSMA) was first proposed as a serum prognostic marker for PCa in 1999; however, its use is controversial [114]. A Dual-Monoclonal Sandwich Assay for PSMA was developed to be used on tissues, seminal fluid and urine [115]. Levels of PSMA in serum have been suggested to be useful for distinguishing between BPH and PCa [116], and subsequently the same results were found for urinary PSMA [117]. PSMA is present in exosomes in urine samples from PCa patients after therapy [118]. Our group has evaluated the utility of PSMA mRNA transcripts in conjunction with PCA3 and Prostate Specific G-coupled Receptor (PSGR) in the PSA diagnostic “gray zone” of 4–10 ng/mL when no prior biopsy information was available. We demonstrated that the prediction of PCa improved significantly for PSMA (0.74), while PSGR (0.66) and PCA3 (0.61) showed a similar performance [119]. However, the use of PSMA has not yet been adopted in clinical practice.
Another promising RNA-based urinary biomarker is encoded by a fusion gene formed as a result of a translocation between the androgen-regulated transmembrane protease, serine 2 (TMPRSS2) gene transcriptional promoter and the ETS related oncogene (ERG), resulting in an androgen-regulated TMPRSS2–ERG fusion gene that is highly specific for PCa and can be found in approximately half of all white PCa patients [120]. Hessels et al., analyzed TMPRSS2-ERG fusion transcripts in urinary sediments and demonstrated a sensitivity of 37% and a specificity of 93% for the prediction of PCa [104]. Moreover, TMPRSS2-ERG was correlated with pathological stage [121], Gleason score [121,122] and with PCa death [122]. Additional marker analysis in a multiplex detection system could further improve sensitivity and specificity.
2.3. miRNA-Based Urine Biomarkers
The discovery of miRNAs has opened up a new field in cancer research with potential novel applications in diagnostics and therapy [123]. MicroRNAs are short, ncRNAs with an average length of 22 nucleotides [124] (see Box II). After transcription they fold into hairpin structures before being processed into mature miRNAs that bind to complementary sequences in mRNAs to alter protein expression. Currently, 1600 precursor and 2042 mature human miRNAs are registered in miRBase Release 19 (August 2012), and each of these may target up to 1000 gene sequences [125]. This provides a complex layer of control in for example, signaling pathways involved in the regulation of cellular functions, ranging from the maintenance of “stemness” to differentiation and tissue development, and from the cell cycle to apoptosis and metabolism [126–128]. Thus, aberrant expression of miRNAs can impact deeply on multiple features of cell biology resulting in complex downstream pathological events, such as cancer [129]. Specific miRNAs have been shown to be abnormally expressed in tumor tissues, playing important roles in cancer onset and disease progression through the targeting of cancer-relevant genes [130].
miRNA profiles of different tissues have been reported to be more predictive than mRNA characterization to such an extent that poorly differentiated tumors of uncertain origin could be classified on the basis of miRNAs expression [131]. MiRNAs are very stable and are detectable in biopsies, serum, and other fluids, such as urine [132]. Between 200 and 500 miRNAs were detected by qPCR in different human body fluids, such as plasma, urine and breast milk [133]. Mitchell et al., found that the serum levels of the miRNA “miR-141” distinguished patients with advanced PCa from healthy controls [134]. Other recent studies have demonstrated that circulating miR-141 levels were correlated to aggressive PCa [135], and that miR-96 and miR-183 expression in urine were well correlated to urothelial carcinoma (UC) stage and grade, serving as promising diagnostic tumor markers capable of distinguishing between UC patients and non-UC patients [136]. However, only one study has been published linking miRNAs from urine with PCa. In that study, the analysis of five selected miRNAs in urine samples found that miR-107 and miR-574-3p were present at a significantly higher concentration in the urine of PCa patients compared to controls [137].
In PCa most of the circulating miRNA studies which have found associations between miRNA populations and aggressive and metastatic disease have been conducted using serum or plasma and need to be validated in larger patient and control samples [130]. Specific miRNA patterns in the urine may also reflect early or advanced PCa disease, but while urine miRNAs have been investigated in bladder and kidney cancer, no comprehensive studies for miRNA in PCa urine have been reported so far. Therefore, despite the obvious potential for circulating and urine miRNAs in diagnostic, prognostic, and predictive applications, clinical implementation of a non-invasive miRNA test for PCa is still a distant goal [138].
2.4. Protein-Based Urine Biomarkers
Protein-based biomarkers include cell-surface receptors, tumor antigens (such as PSA), phosphorylation states, carbohydrate determinants and peptides released by tumors into serum, urine, sputum, nipple aspirates, or other body fluids [109]. Proteins secreted by cancer cells can be essential in the processes of differentiation, invasion and metastasis [143,144]. Secreted proteins or their fragments present in body fluids, such as blood or urine, can be measured via non-invasive or minimally invasive assays. To date, only a few studies have analyzed cancer secretomes. However, the results with regards to the discovery of biomarkers are rather exciting [145].
Recently the detection of under-expressed PSA protein levels in urine has been reported [146–149]. Bolduc et al. compared a small cohort of urine samples collected (without previous DRE) from “normal”, BPH and PCa men, and the data suggested that the ratio of serum PSA to urine PSA could possess diagnostic value [146]. The same idea was also suggested in another independent study where PSA levels were also determined in urine. In that study, no differences between urinary PSA pre- and post-PM were found [150]. Later, Drake et al. [14] performed a study in which they focused on the characterization of PSA and Prostatic Acid Phosphatase (PAP) using an Enzyme-Linked ImmunoSorbent Assay (ELISA) assay on post-DRE urine samples. They found a clear trend towards lower levels of expression for both proteins in their cancer samples.
Another protein-based candidate is Annexin A3 (ANXA3), which is a calcium-binding protein with an associated decreased production in PCa cells. The analysis of ANXA3 using Western blots (WB) of urine samples showed significantly lower values in PCa patients as compared with BPH patients. When this marker was combined with serum PSA there was improved sensitivity and high specificity compared to total PSA, with an AUC of 0.81 [151]. Katafigiotis et al., looked at urine samples from 127 PCa patients obtained after DRE, measuring zinc α 2-glycoprotein (ZAG) by WB. Receiver operating characteristic (ROC) curve analysis showed a significant predictive ability for PCa with AUCs of 0.68 [32].
Recent advances in liquid chromatography (LC) and two dimensional gel electrophoresis (2D-GE), in combination with mass spectrometry (MS) have significantly facilitated the challenging detection of proteins in body fluids [152]. High-throughput proteomic analysis of biological fluids such as urine, has recently become a popular approach for the identification of novel biomarkers, due to the reduced complexity compared to serum [153]. However, only a limited number of studies have focused on PCa.
One of the first proteomic urine profiling experiments for the detection of PCa was performed by Rehman et al., using a gel-based strategy comparing PCa and BPH samples [154]. They identified S100A9 (calgranulin B, MRP-14) as a possible biomarker. However, this data was not verified in an independent study. More recently, several studies have focused on the characterization of urine samples in a high-throughput manner. Teodorescu et al., performed a pilot study for PCa using Capillary Electrophoresis (CE) coupled with MS and to define a potential urinary polypeptide pattern with 92% sensitivity and 96% specificity [155]. Later, the same group described a refinement of the PCa specific biomarker pattern using 51 PCa and 35 BPH urine samples [156]. The model, containing 12 potential biomarkers, resulted in the correct classification of 89% of the PCa cases and 51% of the BPH cases in a second blind cohort of 213 samples. The inclusion of age and free PSA parameters increased the sensitivity and specificity to 91% and 69%, respectively. M’Koma and collaborators performed a large-scale proteomic analysis of BPH, HGPIN and PCa urine samples [157]. Using Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) analysis, the group reported 71.2% specificity and 67.4% sensitivity for discriminating between PCa and BPH, while they also reported a specificity of 73.6% and a sensitivity of 69.2% for discriminating between BPH and HGPIN. Finally, Okamoto et al. used Surface Enhanced Laser Desorption Ionization Time of Flight (SELDI-TOF) analysis coupled to MS to analyze post-DRE urine samples. They obtained a heat map with 72 peaks, which could distinguish PCa from benign lesions with a sensitivity of 91.7% and a specificity of 83.3% [158]. However, although there have been an increasing number of publications in the proteomic urine PCa field, most of this data has not been verified in independent studies.
2.5. Metabolite-Based Urine Biomarkers
Metabolomics is a recently incorporated–omic approach that identifies metabolites using techniques similar to proteomics. Urinary metabolomic profiles have recently drawn a lot of attention owing to a debate regarding their possible role as potential clinical markers for PCa [159]. Using 262 clinical samples, including 110 urine samples, Seekumar et al. performed a major study in the field of PCa metabolomics: 1126 metabolites were analyzed using LC and gas chromatography MS [98], and a profile was identified that was able to distinguish between benign, clinically localized PCa and metastatic cancer. Sarcosine and the N-methyl derivative of the amino acid glycine were found at highly increased levels in PCa and were associated with disease progression to metastasis. However, validation of this metabolite has failed to reproduce these findings [160], and therefore, the utility of sarcosine is still under discussion.
2.6. Urine Biomarker Panels
Although a great number of urine biomarkers have been documented in large screening programs, there are only a few studies that take into account the heterogeneity of cancer development based on a diagnostic profile. Since a single marker may not necessarily reflect the multifactorial nature of PCa, a combination of various biomarkers in conjunction with clinical and demographic data could improve performance over the use of a single biomarker [161–163]. Adding extra genes into the “fingerprint” results in an additional layer of statistical complexity prompting new developments in biostatistics and bioinformatics [109].
Table 2 summarizes the most significant studies that have used panels of urinary biomarkers. Hessels et al. performed a study on 108 patients using urine sediments, where the authors combined PCA3 with TMPRSS2-ERG fusion status. Combining both markers remarkably increased the sensitivity for the detection of PCa [104]. In this sense, the combination of TMPRSS2-ERG and PCA3 and serum PSA was described as a method that could predict PCa with 80% sensitivity and 90% specificity [161] and help urologists in the decision to take PBs [162]. Furthermore, TMPRSS2-ERG in combination with PCA3 enhances serum PSA as a marker for defining PCa risk and clinically relevant cancer on PB [163]. More recently, Lin and collaborators also combined these markers and demonstrated that they can be used to stratify the risk of having aggressive PCa [54]. Another important study came from Lexman et al., who developed a multiplex model that measured the expression of seven putative PCa biomarkers and found that a combination of Golgi Membrane Protein (GOLPH2), Serine Peptidase Inhibitor Kazal type 1 (SPINK1) and PCA3 transcript expression with TMPRSS2-ERG fusion status was a better predictor of PCa than PSA or PCA3 alone (65.9% sensitivity and 76.0% specificity) [54]. Ouyang et al., have developed a duplex qPCR assay for the detection of PCa, based on the quantification of alpha-methylacyl-CoA racemase (AMACR) and PCA3 in urine sediments, while Talesa et al. analyzed PSMA, Hepsin (HPN), PCA3, UDP-n-acetyl-alpha-d-galactosamine: polypeptide N-acetylgalactosaminyltransferase 3 (GalNAC-T3) and PSA using qPCR and concluded that the best combination of biomarkers for predictors of PCa included urinary PSA and PSMA [117].
Table 2.
Summary of the most significant studies that have used panels of urine biomarkers for PCa detection.
| Biomarker type | Study | Marker | PCa/study | Sens. | Spec. | AUC |
|---|---|---|---|---|---|---|
| DNA | Hoque et al., 2005 [112] | p16, ARF, MGMT, GSTP1 | 73 | 87% | 100% | ND |
| Rouprêt et al., 2007 [110] | GSTP1, RASSF1A, RARB, and APC | 95/133 | 87% | 89% | ND | |
| Vener et al., 2008 [165] | GSTP1, RARB and APC | 54/121 | 55% | 80% | 0.69 | |
| Payne et al., 2009 [166] | GSTP1, RASSF2, HIS1H4K, TFAP2E | 192 | 94% | 27% | ||
| Baden et al., 2009 [164] | GSTP1, RARB and APC | 178/159 | ND | ND | 0.72 | |
| Costa et al., 2011 [113] | PCDH17, TCF21 | 318 | 26% | 100% | ||
|
| ||||||
| mRNA | Hessels et al., 2007 [104] | PCA3 and TMPRSS2:ERG | 78/108 | 73% | 52% | ND |
|
| ||||||
| Laxman et al., 2008 [54] | PCA3, GOLPH2, SPINK1 and TMPRSS2:ERG | 152/257 | 66% | 76% | 0.76 | |
|
| ||||||
| Ouyang et al., 2009 [167] | AMACR and PCA3 | 43/92 | 72% | 53% | ND | |
|
| ||||||
| Talesa et al., 2009 [117] | PSMA, HPN, PCA3, GalNAC-T3 and serum PSA | 49% | ND | ND | ||
|
| ||||||
| Rigau et al., 2010 [81] | PCA3 and PSGR | 73/215 | 96% | 34% | 0.73 | |
|
| ||||||
| Rigau et al., 2011 [119] | PSMA, PSGR, PCA3 and serum PSA | 57/154 | 96% | 50% | 0.82 | |
|
| ||||||
| Salami et al., 2011 [168] | PCA3, TMPRSS2:ERG and serum PSA | 15/45 | 80% | 90% | 0.88 | |
|
| ||||||
| Jamasphvili et al., 2011 [169] | PCA3, AMACR, TRMP8, SMSB | 104 | 72% | 71% | ||
| Nguyen et al., 2011 [170] | TMPRSS2:ERG subtypes | 101 | 35% | 100% | ||
|
| ||||||
| Tomlins et al., 2011 [171] | PCA3 and TMPRSS2:ERG | 463 (acad.) and 439 (biopsy) | a_0.64 and b_0.66 | |||
|
| ||||||
| Protein | Rehman et al., 2004 [154] | ENO1, IDH3B, B2M, A1M, PRO2044 and S100A9 (Calgranulin_B/MRP-14) | 6 PC (12) | |||
|
| ||||||
| Theodorescu et al., 2005 [155] | Proteinpolypeptide | 26/47 | 92% | 96% | ||
|
| ||||||
| M’Koma et al., 2007 [157] | 130 m/z | 89/407 | 81% | 80% | ||
|
| ||||||
| Theodorescu et al., 2008 [156] | 12 protein pannel + age + serum PSA | 86 Training set + 213 validation set | 91% | 62% | ||
|
| ||||||
| Okamoto et al., 2008 [158] | 72 masspicks | 57/113 | 91% | 83% | ||
|
| ||||||
| Mixture | Cao et al., 2010 [172] | mRNA, protein and metabolite (PCA3, TMPRSS2: ERG, ANXA3, Sarcosine, and urine PSA) | 86/131 | 95% | 50% | 0.86 |
|
| ||||||
| Prior et al., 2010 [173] | mRNA (AMACR/MMP2), DNA (GSTP1/RASSF1A) and PSA in serum and urine | 34/113 | 57% | 97% | 0.79 | |
Rigau et al. [119] have developed a multiplex test based on the combination of qPCR analysis of PCA3, PSGR, PSMA levels in urine with serum PSA protein levels in a prospective study using post DRE urine samples from 57 PCa patients and 97 age-matched benign controls. They observed that by using this model, it is possible to reduce the number of unnecessary PB by 34% [119]. A multiplexed quantitative methylation-specific PCR assay consisting of three different methylated genes: GSTP1, RARB and APC was recently tested in a prospective multicenter study using post-DRE urine samples from 178 PCa patients and 159 controls. The predictive accuracy AUC of the assay for detecting PCa was 0.72. This was only a marginal gain in predictive ability with respect to biopsy outcome as compared to total PSA and DRE alone [164]. Although these combined biomarkers significantly improve sensitivity and specificity over single biomarkers, to our knowledge none of these panels have yet been established in clinical practice.
2.7. Exosomes as a Source of Urine Biomarkers
Exosomes are small, secreted membranous vesicles formed in multivesicular bodies through an inward budding mechanism that encapsulates cytoplasmic components [174]. For many years exosomes were thought to be organelles for the removal of cell debris or obsolete surface molecules from the cell. However, further investigations have revealed a role for exosomes in inter-cellular communication. In the last five years, several studies have demonstrated that exosomes may be secreted by multiple cell lines and cell types, including tumor cell lines, stem cells and neuronal cells [175]. In addition, exosomes have been identified in most body fluids, such as blood, urine and ascites [175]. The discovery of their nucleic acid contents, such as mRNA, small ncRNA, miRNA and mitochondrial DNA (mtDNA), which can be transported to other cells [176], represents a major breakthrough, and several studies have indicated that they can play a novel role as regulators in cell-cell communication during diverse biological processes. Urinary exosomes have recently been described as treasure chests of information and a potential source of new cancer biomarkers including PCa [15]. Analyzing the content of exosomes harvested from urine has a number of advantages: (i) it is non-invasive; (ii) data is informative with regards to PCa diagnosis and potentially the status of overall tumor malignancy; (iii) the genetic and proteomic material within exosomes is protected from enzymic degradation by the exosomal lipid bilayer [177], and (iv) exosomes are stable after long-term storage at −80 °C, which makes prospective studies feasible. Further progress has been made in terms of storage, processing [178] and analysis of protein [22] and RNA content.
To our knowledge no high-throughput technique has been used to analyze the RNA or protein content of urinary exosomes for PCa biomarker discovery in individual samples. However, some reports have indicated urinary exosomes to be an excellent source of PCa biomarkers. At a protein level, Mitchell PJ et al. [118] analyzed urinary exosomes from 10 healthy donors and 10 PCa patients who were undergoing hormonal therapy prior to radical radiotherapy. PSA and PSMA were found to be present in almost all of the PCa specimens, but not in the healthy donor specimens. At an RNA level, Nilsson et al. [179] showed that known RNA markers for PCa, such as TMPRSS2-ERG fusion transcripts and PCA3, could be detected in urine-derived and PCa cell line-derived exosomes by using Nested PCR [24]. This demonstrated a potential for diagnosis, as well as a strategy for the successful monitoring of the status of cancer patients. miRNAs have also been detected in extracellular fractions, stabilised by their encapsulation in microvesicles such as exosomes. Exosomes are thus a prime non-invasive source of biomarkers for cancer and other diseases [180].
3. Conclusions
The introduction of PSA testing has radically altered how PCa is diagnosed and managed. However, controversy still exists regarding both the utility of PSA screening for reducing PCa mortality and the risks associated with PCa over-diagnosis. Furthermore, there is the problem of the heterogeneous nature of PCa foci and problem of adequately sampling and assessing foci of poor prognosis tumor. Additional markers are therefor urgently required to supplement or replace the PSA test and improve the specificity of PCa detection and prognosis. Multiplex urine-based assays could provide the answer and have the advantage of potentially sampling PCa material from multiple tumor foci within individual prostates and providing both diagnostic and prognostic biomarkers [181].
It has been demonstrated that post-DRE urine samples are a rich source of biomarkers for PCa. Urine can be obtained in any urology clinic and does not require any change in routine clinical practices. Thus, post-DRE urine could be the best compromise between a minimally invasive technique and obtaining sufficient material for a correct diagnosis. However, to properly assess and validate promising urine candidates there needs to be large prospective studies of urine biomarkers using robust and standardized methods for urine collection, storage, harvest and analysis of DNA, RNA, miRNA, protein and metabolites.
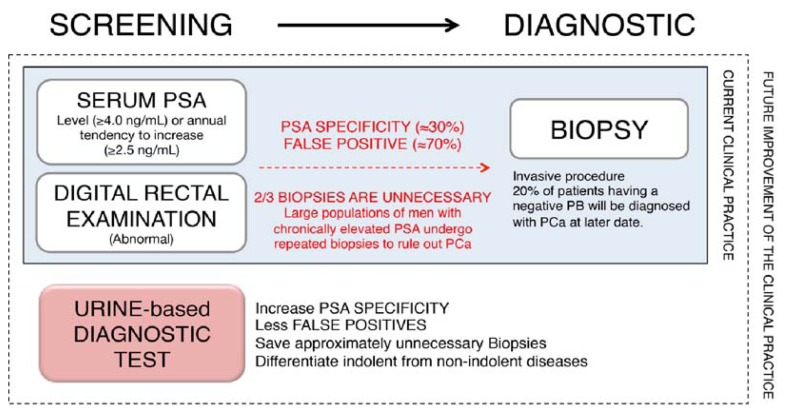
A future goal is therefore the development of a low cost, point of care, multiplexed, urine-based detection test for PCa which could be incorporated seamlessly into routine clinical practice to better determine which patients should undergo biopsy, and to highlight those patients that have a high risk of PCa metastasis/CRPC, and which therefor require treatment, at the earliest possible point in time (Figure 2).
Figure 2.
Current and future improvement in the PCa diagnostic scheme.
In summary, the future of urine-based PCa biomarkers looks promising. It remains for us to validate the many exciting candidate biomarkers that have been discovered and to discover novel markers that will help to: (i) identify those men with indolent PCa, i.e., those who will not be affected by disease in their lifetimes and who do not need treatment; (ii) minimize the number of unnecessary PBs; (iii) identify men with aggressive disease, distinguishing between who will benefit from local therapy and those who are likely to fail local therapy and require adjuvant intervention; and (iv) find markers that may serve as surrogate end points for clinical progression or survival [182].
Another important point that needs to be addressed is the necessity of the DRE. In the future, we would like to know if urine samples provided without a DRE contain enough material to correctly detect prostate biomarkers and, thus, enable a correct diagnosis. Although DRE is part of the diagnostic tripod (PSA, DRE and biopsy), it is usually poorly tolerated by patients and always requires medical intervention. This detail may represent a limiting factor, since the urologist would need to have the facilities to freeze and store urine samples before sending them to the laboratory. In large trials, the question of whether and how to perform the DRE to optimize sensitivity and specificity must be addressed for each potential marker [183].
Acknowledgments
Instituto de Salud Carlos III: PI11/02486, CP10/00355, PS09/00496, Ministerio de Ciéncia e Innovación: RTICC RD06/0020/0058; Asociación Española Contra el Cáncer Junta Provincial de Barcelona; Red de Genómica del Cáncer y Genotipado de tumores C03/10; Fundación para la Investigación en Urología, Departament d’Univeristats, Recerca i Societat de la Informació de la Generalitat de Catalunya: SGR00487; Movember Foundation 2012, Valor 2010/00220, programa INNPACTO and Lisa Piccione for reviewing the document.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Parkin D.M., Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur. J. Cancer. 2010;46:765–781. doi: 10.1016/j.ejca.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Strope S.A., Andriole G.L. Prostate cancer screening: Current status and future perspectives. Nat. Rev. Urol. 2010;7:487–493. doi: 10.1038/nrurol.2010.120. [DOI] [PubMed] [Google Scholar]
- 4.Bretton P.R. Prostate-specific antigen and digital rectal examination in screening for prostate cancer: A community-based study. South Med. J. 1994;87:720–723. doi: 10.1097/00007611-199407000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Catalona W.J., Richie J.P., Ahmann F.R., Hudson M.A., Scardino P.T., Flanigan R.C., deKernion J.B., Ratliff T.L., Kavoussi L.R., Dalkin B.L., et al. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: Results of a multicenter clinical trial of 6630 men. J. Urol. 1994;151:1283–1290. doi: 10.1016/s0022-5347(17)35233-3. [DOI] [PubMed] [Google Scholar]
- 6.Catalona W.J., Smith D.S., Ratliff T.L., Dodds K.M., Coplen D.E., Yuan J.J., Petros J.A., Andriole G.L. Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N. Engl. J. Med. 1991;324:1156–1161. doi: 10.1056/NEJM199104253241702. [DOI] [PubMed] [Google Scholar]
- 7.Tuma R.S. New tests for prostate cancer may be nearing the clinic. J. Natl. Cancer Inst. 2010;102:752–754. doi: 10.1093/jnci/djq210. [DOI] [PubMed] [Google Scholar]
- 8.Thompson I.M., Ankerst D.P., Chi C., Lucia M.S., Goodman P.J., Crowley J.J., Parnes H.L., Coltman C.A., Jr Operating characteristics of prostate-specific antigen in men with an initial PSA level of 3.0 ng/mL or lower. JAMA. 2005;294:66–70. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 9.Thompson I.M., Pauler D.K., Goodman P.J., Tangen C.M., Lucia M.S., Parnes H.L., Minasian L.M., Ford L.G., Lippman S.M., Crawford E.D., et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or =4.0 ng per milliliter. N. Engl. J. Med. 2004;350:2239–2246. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 10.Schroder F.H., van der Cruijsen-Koeter I., de Koning H.J., Vis A.N., Hoedemaeker R.F., Kranse R. Prostate cancer detection at low prostate specific antigen. J. Urol. 2000;163:806–812. [PubMed] [Google Scholar]
- 11.Gerszten R.E., Wang T.J. The search for new cardiovascular biomarkers. Nature. 2008;451:949–952. doi: 10.1038/nature06802. [DOI] [PubMed] [Google Scholar]
- 12.Boja E., Hiltke T., Rivers R., Kinsinger C., Rahbar A., Mesri M., Rodriguez H. Evolution of Clinical Proteomics and its Role in Medicine. J. Proteome Res. 2010;10:66–84. doi: 10.1021/pr100532g. [DOI] [PubMed] [Google Scholar]
- 13.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J. Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake R.R., White K.Y., Fuller T.W., Igwe E., Clements M.A., Nyalwidhe J.O., Given R.W., Lance R.S., Semmes O.J. Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease. J. Proteomics. 2009;72:907–917. doi: 10.1016/j.jprot.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duijvesz D., Luider T., Bangma C.H., Jenster G. Exosomes as biomarker treasure chests for prostate cancer. Eur. Urol. 2011;59:823–831. doi: 10.1016/j.eururo.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 16.Foot N.C., Papanicolaou G.N., Holmquist N.D., Seybolt J.F. Exfoliative cytology of urinary sediments; a review of 2829 cases. Cancer. 1958;11:127–137. doi: 10.1002/1097-0142(195801/02)11:1<127::aid-cncr2820110124>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan B., Truong L.D. Prostatic adenocarcinoma diagnosed by urinary cytology. Am. J. Clin. Pathol. 2000;113:29–34. doi: 10.1309/4t6h-549r-capj-fey0. [DOI] [PubMed] [Google Scholar]
- 18.Rifai N., Gillette M.A., Carr S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 19.You J., Cozzi P., Walsh B., Willcox M., Kearsley J., Russell P., Li Y. Innovative biomarkers for prostate cancer early diagnosis and progression. Crit. Rev. Oncol. Hematol. 2010;73:10–22. doi: 10.1016/j.critrevonc.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Zehentner B.K., Secrist H., Zhang X., Hayes D.C., Ostenson R., Goodman G., Xu J., Kiviat M., Kiviat N., Persing D.H., et al. Detection of alpha-methylacyl-coenzyme-A racemase transcripts in blood and urine samples of prostate cancer patients. Mol. Diagn Ther. 2006;10:397–403. doi: 10.1007/BF03256217. [DOI] [PubMed] [Google Scholar]
- 21.Sreekumar A., Laxman B., Rhodes D.R., Bhagavathula S., Harwood J., Giacherio D., Ghosh D., Sanda M.G., Rubin M.A., Chinnaiyan A.M. Humoral immune response to alpha-methylacyl-CoA racemase and prostate cancer. J. Natl. Cancer Inst. 2004;9(6):834–843. doi: 10.1093/jnci/djh145. [DOI] [PubMed] [Google Scholar]
- 22.Pisitkun T., Shen R.F., Knepper M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wozny W., Schroer K., Schwall G.P., Poznanovic S., Stegmann W., Dietz K., Rogatsch H., Schaefer G., Huebl H., Klocker H., et al. Differential radioactive quantification of protein abundance ratios between benign and malignant prostate tissues: Cancer association of annexin A3. Proteomics. 2007;7:313–322. doi: 10.1002/pmic.200600646. [DOI] [PubMed] [Google Scholar]
- 24.Gerke V., Creutz C.E., Moss S.E. Annexins: Linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y., Li J., Yu X., Li S., Zhang X., Mo Z., Hu Y. APC gene hypermethylation and prostate cancer: A systematic review and meta-analysis. Eur. J. Hum. Genet. 2013 doi: 10.1038/ejhg.2012.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foley R., Marignol L., Keane J.P., Lynch T.H., Hollywood D. Androgen hypersensitivity in prostate cancer: Molecular perspectives on androgen deprivation therapy strategies. Prostate. 2010;71:550–557. doi: 10.1002/pros.21266. [DOI] [PubMed] [Google Scholar]
- 27.Massie C.E., Lynch A., Ramos-Montoya A., Boren J., Stark R., Fazli L., Warren A., Scott H., Madhu B., Sharma N., et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu R., Isaacs W.B., Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656–1667. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorlov I.P., Sircar K., Zhao H., Maity S.N., Navone N.M., Gorlova O.Y., Troncoso P., Pettaway C.A., Byun J.Y., Logothetis C.J. Prioritizing genes associated with prostate cancer development. BMC Cancer. 2010;10:599. doi: 10.1186/1471-2407-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aparicio A., Logothetis C.J., Maity S.N. Understanding the lethal variant of prostate cancer: Power of examining extremes. Cancer Discov. 2011;1:466–468. doi: 10.1158/2159-8290.CD-11-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosquera J.M., Beltran H., Park K., MacDonald T.Y., Robinson B.D., Tagawa S.T., Perner S., Bismar T.A., Erbersdobler A., Dhir R., et al. Concurrent AURKA and MYCN gene amplifications are harbingers of lethal treatment-related neuroendocrine prostate cancer. Neoplasia. 2013;15:1–10. doi: 10.1593/neo.121550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katafigiotis I., Tyritzis S.I., Stravodimos K.G., Alamanis C., Pavlakis K., Vlahou A., Makridakis M., Katafigioti A., Garbis S.D., Constantinides C.A. Zinc alpha2-glycoprotein as a potential novel urine biomarker for the early diagnosis of prostate cancer. BJU Int. 2012;11:688–693. doi: 10.1111/j.1464-410X.2012.11501.x. [DOI] [PubMed] [Google Scholar]
- 33.Yip P.Y., Kench J.G., Rasiah K.K., Benito R.P., Lee C.S., Stricker P.D., Henshall S.M., Sutherland R.L., Horvath L.G. Low AZGP1 expression predicts for recurrence in margin-positive, localized prostate cancer. Prostate. 2011;71:1638–1645. doi: 10.1002/pros.21381. [DOI] [PubMed] [Google Scholar]
- 34.Bondar O.P., Barnidge D.R., Klee E.W., Davis B.J., Klee G.G. LC-MS/MS quantification of Zn-alpha2 glycoprotein: A potential serum biomarker for prostate cancer. Clin. Chem. 2007;53:673–678. doi: 10.1373/clinchem.2006.079681. [DOI] [PubMed] [Google Scholar]
- 35.Palanisamy N., Ateeq B., Kalyana-Sundaram S., Pflueger D., Ramnarayanan K., Shankar S., Han B., Cao Q., Cao X., Suleman K., et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat. Med. 2010;16:793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J., Kobayashi T., Floc’h N., Kinkade C.W., Aytes A., Dankort D., Lefebvre C., Mitrofanova A., Cardiff R.D., McMahon M., et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer Res. 2012;72:4765–4776. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren G., Liu X., Mao X., Zhang Y., Stankiewicz E., Hylands L., Song R., Berney D.M., Clark J., Cooper C., et al. Identification of frequent BRAF copy number gain and alterations of RAF genes in Chinese prostate cancer. Genes Chromosomes Cancer. 2012;51:1014–1023. doi: 10.1002/gcc.21984. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K., Furihata M., Tsunoda T., Ashida S., Takata R., Obara W., Yoshioka H., Daigo Y., Nasu Y., Kumon H., et al. Molecular features of hormone-refractory prostate cancer cells by genome-wide gene expression profiles. Cancer Res. 2007;67:5117–5125. doi: 10.1158/0008-5472.CAN-06-4040. [DOI] [PubMed] [Google Scholar]
- 39.Karacosta L.G., Foster B.A., Azabdaftari G., Feliciano D.M., Edelman A.M. A regulatory feedback loop between Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) and the androgen receptor in prostate cancer progression. J. Biol. Chem. 2012;287:24832–24843. doi: 10.1074/jbc.M112.370783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima T., Mizokami A., Miyagi T., Kawai K., Izumi K., Kumaki M., Ofude M., Zhang J., Keller E.T., Namiki M. Down-regulation of calcium/calmodulin-dependent protein kinase kinase 2 by androgen deprivation induces castration-resistant prostate cancer. Prostate. 2012;72:1789–1801. doi: 10.1002/pros.22533. [DOI] [PubMed] [Google Scholar]
- 41.Umbas R., Isaacs W.B., Bringuier P.P., Schaafsma H.E., Karthaus H.F., Oosterhof G.O., Debruyne F.M., Schalken J.A. Decreased E-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res. 1994;54:3929–3933. [PubMed] [Google Scholar]
- 42.Umbas R., Schalken J.A., Aalders T.W., Carter B.S., Karthaus H.F., Schaafsma H.E., Debruyne F.M., Isaacs W.B. Expression of the cellular adhesion molecule E-cadherin is reduced or absent in high-grade prostate cancer. Cancer Res. 1992;52:5104–5109. [PubMed] [Google Scholar]
- 43.Chi K.N., Zoubeidi A., Gleave M.E. Custirsen (OGX-011): A second-generation antisense inhibitor of clusterin for the treatment of cancer. Expert Opin. Investig Drugs. 2008;17:1955–1962. doi: 10.1517/13543780802528609. [DOI] [PubMed] [Google Scholar]
- 44.Chen M., Wang K., Zhang L., Li C., Yang Y. The discovery of putative urine markers for the specific detection of prostate tumor by integrative mining of public genomic profiles. PLoS One. 2011;6:e28552. doi: 10.1371/journal.pone.0028552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosseini-Beheshti E., Pham S., Adomat H., Li N., Tomlinson Guns E.S. Exosomes as biomarker enriched microvesicles: Characterization of exosomal proteins derived from a panel of prostate cell lines with distinct AR phenotypes. Mol. Cell Proteomics. 2011;11:863–885. doi: 10.1074/mcp.M111.014845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamoureux F., Thomas C., Yin M.J., Kuruma H., Beraldi E., Fazli L., Zoubeidi A., Gleave M.E. Clusterin inhibition using OGX-011 synergistically enhances Hsp90 inhibitor activity by suppressing the heat shock response in castrate-resistant prostate cancer. Cancer Res. 2011;71:5838–5849. doi: 10.1158/0008-5472.CAN-11-0994. [DOI] [PubMed] [Google Scholar]
- 47.Bjartell A.S., Al-Ahmadie H., Serio A.M., Eastham J.A., Eggener S.E., Fine S.W., Udby L., Gerald W.L., Vickers A.J., Lilja H., et al. Association of cysteine-rich secretory protein 3 and beta-microseminoprotein with outcome after radical prostatectomy. Clin. Cancer Res. 2007;13:4130–4138. doi: 10.1158/1078-0432.CCR-06-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bjartell A., Johansson R., Bjork T., Gadaleanu V., Lundwall A., Lilja H., Kjeldsen L., Udby L. Immunohistochemical detection of cysteine-rich secretory protein 3 in tissue and in serum from men with cancer or benign enlargement of the prostate gland. Prostate. 2006;66:591–603. doi: 10.1002/pros.20342. [DOI] [PubMed] [Google Scholar]
- 49.Dhir R., Vietmeier B., Arlotti J., Acquafondata M., Landsittel D., Masterson R., Getzenberg R.H. Early identification of individuals with prostate cancer in negative biopsies. J. Urol. 2004;171:1419–1423. doi: 10.1097/01.ju.0000116545.94813.27. [DOI] [PubMed] [Google Scholar]
- 50.Paul B., Dhir R., Landsittel D., Hitchens M.R., Getzenberg R.H. Detection of prostate cancer with a blood-based assay for early prostate cancer antigen. Cancer Res. 2005;65:4097–4100. doi: 10.1158/0008-5472.CAN-04-4523. [DOI] [PubMed] [Google Scholar]
- 51.Leman E.S., Cannon G.W., Trock B.J., Sokoll L.J., Chan D.W., Mangold L., Partin A.W., Getzenberg R.H. EPCA-2: A highly specific serum marker for prostate cancer. Urology. 2007;69:714–720. doi: 10.1016/j.urology.2007.01.097. [DOI] [PubMed] [Google Scholar]
- 52.Horoszewicz J.S., Kawinski E., Murphy G.P. Monoclonal antibodies to a new antigenic marker in epithelial prostatic cells and serum of prostatic cancer patients. Anticancer Res. 1987;7:927–935. [PubMed] [Google Scholar]
- 53.Zhang Y., Guo Z., Du T., Chen J., Wang W., Xu K., Lin T., Huang H. Prostate specific membrane antigen (PSMA): A novel modulator of p38 for proliferation, migration, and survival in prostate cancer cells. Prostate. 2012;73:835–841. doi: 10.1002/pros.22627. [DOI] [PubMed] [Google Scholar]
- 54.Laxman B., Morris D.S., Yu J., Siddiqui J., Cao J., Mehra R., Lonigro R.J., Tsodikov A., Wei J.T., Tomlins S.A., et al. A first-generation multiplex biomarker analysis of urine for the early detection of prostate cancer. Cancer Res. 2008;68:645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Varambally S., Laxman B., Mehra R., Cao Q., Dhanasekaran S.M., Tomlins S.A., Granger J., Vellaichamy A., Sreekumar A., Yu J., et al. Golgi protein GOLM1 is a tissue and urine biomarker of prostate cancer. Neoplasia. 2008;10:1285–1294. doi: 10.1593/neo.08922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonzalgo M.L., Nakayama M., Lee S.M., de Marzo A.M., Nelson W.G. Detection of GSTP1 methylation in prostatic secretions using combinatorial MSP analysis. Urology. 2004;63:414–418. doi: 10.1016/j.urology.2003.08.039. [DOI] [PubMed] [Google Scholar]
- 57.Crocitto L.E., Korns D., Kretzner L., Shevchuk T., Blair S.L., Wilson T.G., Ramin S.A., Kawachi M.H., Smith S.S. Prostate cancer molecular markers GSTP1 and hTERT in expressed prostatic secretions as predictors of biopsy results. Urology. 2004;64:821–825. doi: 10.1016/j.urology.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Stephan C., Yousef G.M., Scorilas A., Jung K., Jung M., Kristiansen G., Hauptmann S., Kishi T., Nakamura T., Loening S.A., et al. Hepsin is highly over expressed in and a new candidate for a prognostic indicator in prostate cancer. J. Urol. 2004;17(1):187–191. doi: 10.1097/01.ju.0000101622.74236.94. [DOI] [PubMed] [Google Scholar]
- 59.Dhanasekaran S.M., Barrette T.R., Ghosh D., Shah R., Varambally S., Kurachi K., Pienta K.J., Rubin M.A., Chinnaiyan A.M. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 60.Shariat S.F., Andrews B., Kattan M.W., Kim J., Wheeler T.M., Slawin K.M. Plasma levels of interleukin-6 and its soluble receptor are associated with prostate cancer progression and metastasis. Urology. 2001;58:1008–1015. doi: 10.1016/s0090-4295(01)01405-4. [DOI] [PubMed] [Google Scholar]
- 61.Nakashima J., Tachibana M., Horiguchi Y., Oya M., Ohigashi T., Asakura H., Murai M. Serum interleukin 6 as a prognostic factor in patients with prostate cancer. Clin. Cancer Res. 2000;6:2702–2706. [PubMed] [Google Scholar]
- 62.Shariat S.F., Kattan M.W., Traxel E., Andrews B., Zhu K., Wheeler T.M., Slawin K.M. Association of pre- and postoperative plasma levels of transforming growth factor beta(1) and interleukin 6 and its soluble receptor with prostate cancer progression. Clin. Cancer Res. 2004;10:1992–1999. doi: 10.1158/1078-0432.ccr-0768-03. [DOI] [PubMed] [Google Scholar]
- 63.Han Z.D., Zhang Y.Q., He H.C., Dai Q.S., Qin G.Q., Chen J.H., Cai C., Fu X., Bi X.C., Zhu J.G., et al. Identification of novel serological tumor markers for human prostate cancer using integrative transcriptome and proteome analysis. Med. Oncol. 2012;29:2877–2888. doi: 10.1007/s12032-011-0149-9. [DOI] [PubMed] [Google Scholar]
- 64.Darson M.F., Pacelli A., Roche P., Rittenhouse H.G., Wolfert R.L., Young C.Y., Klee G.G., Tindall D.J., Bostwick D.G. Human glandular kallikrein 2 (hK2) expression in prostatic intraepithelial neoplasia and adenocarcinoma: A novel prostate cancer marker. Urology. 1997;49:857–862. doi: 10.1016/s0090-4295(97)00108-8. [DOI] [PubMed] [Google Scholar]
- 65.Haese A., Graefen M., Steuber T., Becker C., Pettersson K., Piironen T., Noldus J., Huland H., Lilja H. Human glandular kallikrein 2 levels in serum for discrimination of pathologically organ-confined from locally-advanced prostate cancer in total PSA-levels below 10 ng/mL. Prostate. 2001;49:101–109. doi: 10.1002/pros.1123. [DOI] [PubMed] [Google Scholar]
- 66.Aprikian A. PSA for prostate cancer detection: In serum, in urine or both? Can. Urol. Assoc. J. 2007;1:382. doi: 10.5489/cuaj.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W., Mize G.J., Zhang X., Takayama T.K. Kallikrein-related peptidase-4 initiates tumor-stroma interactions in prostate cancer through protease-activated receptor-1. Int. J. Cancer. 2010;126:599–610. doi: 10.1002/ijc.24904. [DOI] [PubMed] [Google Scholar]
- 68.Avgeris M., Stravodimos K., Scorilas A. Kallikrein-related peptidase 4 gene (KLK4) in prostate tumors: Quantitative expression analysis and evaluation of its clinical significance. Prostate. 2011;71:1780–1789. doi: 10.1002/pros.21395. [DOI] [PubMed] [Google Scholar]
- 69.Pressinotti N.C., Klocker H., Schafer G., Luu V.D., Ruschhaupt M., Kuner R., Steiner E., Poustka A., Bartsch G., Sultmann H. Differential expression of apoptotic genes PDIA3 and MAP3K5 distinguishes between low- and high-risk prostate cancer. Mol. Cancer. 2009;8:130. doi: 10.1186/1476-4598-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bubendorf L., Tapia C., Gasser T.C., Casella R., Grunder B., Moch H., Mihatsch M.J., Sauter G. Ki67 labeling index in core needle biopsies independently predicts tumor-specific survival in prostate cancer. Hum. Pathol. 1998;29:949–954. doi: 10.1016/s0046-8177(98)90199-x. [DOI] [PubMed] [Google Scholar]
- 71.Cuzick J., Swanson G.P., Fisher G., Brothman A.R., Berney D.M., Reid J.E., Mesher D., Speights V.O., Stankiewicz E., Foster C.S., et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2009;12:245–255. doi: 10.1016/S1470-2045(10)70295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zellweger T., Gunther S., Zlobec I., Savic S., Sauter G., Moch H., Mattarelli G., Eichenberger T., Curschellas E., Rufenacht H., et al. Tumor growth fraction measured by immunohistochemical staining of Ki67 is an independent prognostic factor in preoperative prostate biopsies with small-volume or low-grade prostate cancer. Int. J. Cancer. 2009;124:2116–2123. doi: 10.1002/ijc.24174. [DOI] [PubMed] [Google Scholar]
- 73.Riddick A.C., Shukla C.J., Pennington C.J., Bass R., Nuttall R.K., Hogan A., Sethia K.K., Ellis V., Collins A.T., Maitland N.J., et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br. J. Cancer. 2005;92:2171–2180. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee S., Desai K.K., Iczkowski K.A., Newcomer R.G., Wu K.J., Zhao Y.G., Tan W.W., Roycik M.D., Sang Q.X. Coordinated peak expression of MMP-26 and TIMP-4 in preinvasive human prostate tumor. Cell Res. 2006;16:750–758. doi: 10.1038/sj.cr.7310089. [DOI] [PubMed] [Google Scholar]
- 75.Shukla C.J., Pennington C.J., Riddick A.C., Sethia K.K., Ball R.Y., Edwards D.R. Laser-capture microdissection in prostate cancer research: Establishment and validation of a powerful tool for the assessment of tumor-stroma interactions. BJU Int. 2008;101:765–74. doi: 10.1111/j.1464-410X.2007.07372.x. [DOI] [PubMed] [Google Scholar]
- 76.Zhao Y.G., Xiao A.Z., Ni J., Man Y.G., Sang Q.X. Expression of matrix metalloproteinase-26 in multiple human cancer tissues and smooth muscle cells. Ai Zheng. 2009;28:1168–1175. doi: 10.5732/cjc.008.10768. [DOI] [PubMed] [Google Scholar]
- 77.Moses M.A., Wiederschain D., Loughlin K.R., Zurakowski D., Lamb C.C., Freeman M.R. Increased incidence of matrix metalloproteinases in urine of cancer patients. Cancer Res. 1998;58:1395–1399. [PubMed] [Google Scholar]
- 78.Roy R., Louis G., Loughlin K.R., Wiederschain D., Kilroy S.M., Lamb C.C., Zurakowski D., Moses M.A. Tumor-specific urinary matrix metalloproteinase fingerprinting: Identification of high molecular weight urinary matrix metalloproteinase species. Clin. Cancer Res. 2008;14:6610–6617. doi: 10.1158/1078-0432.CCR-08-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu L.L., Stackhouse B.G., Florence K., Zhang W., Shanmugam N., Sesterhenn I.A., Zou Z., Srikantan V., Augustus M., Roschke V., et al. PSGR, a novel prostate-specific gene with homology to a G protein-coupled receptor, is overexpressed in prostate cancer. Cancer Res. 2000;60:6568–6572. [PubMed] [Google Scholar]
- 80.Xu L.L., Sun C., Petrovics G., Makarem M., Furusato B., Zhang W., Sesterhenn I.A., McLeod D.G., Sun L., Moul J.W., et al. Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:56–61. doi: 10.1038/sj.pcan.4500836. [DOI] [PubMed] [Google Scholar]
- 81.Rigau M., Morote J., Mir M.C., Ballesteros C., Ortega I., Sanchez A., Colas E., Garcia M., Ruiz A., Abal M., et al. PSGR and PCA3 as biomarkers for the detection of prostate cancer in urine. Prostate. 2010;70:1760–1767. doi: 10.1002/pros.21211. [DOI] [PubMed] [Google Scholar]
- 82.Gutman A.B. The development of the acid phosphatase test for prostatic carcinoma: The Sixth Ferdinand C. Valentine Memorial Lecture. Bull. N. Y. Acad. Med. 1968;44:63–76. [PMC free article] [PubMed] [Google Scholar]
- 83.Kim Y., Ignatchenko V., Yao C.Q., Kalatskaya I., Nyalwidhe J.O., Lance R.S., Gramolini A.O., Troyer D.A., Stein L.D., Boutros P.C., et al. Identification of differentially expressed proteins in direct expressed prostatic secretions of men with organ-confined versus extracapsular prostate cancer. Mol. Cell Proteomics. 2012;11:1870–1884. doi: 10.1074/mcp.M112.017889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leyten G.H., Hessels D., Jannink S.A., Smit F.P., de Jong H., Cornel E.B., de Reijke T.M., Vergunst H., Kil P., Knipscheer B.C., et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur. Urol. 2012 doi: 10.1016/j.eururo.2012.11.014. [DOI] [PubMed] [Google Scholar]
- 85.Bussemakers M.J., van Bokhoven A., Verhaegh G.W., Smit F.P., Karthaus H.F., Schalken J.A., Debruyne F.M., Ru N., Isaacs W.B. DD3: A new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;5(9):5975–5979. [PubMed] [Google Scholar]
- 86.Day J.R., Jost M., Reynolds M.A., Groskopf J., Rittenhouse H. PCA3: From basic molecular science to the clinical lab. Cancer Lett. 2011;301:1–6. doi: 10.1016/j.canlet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 87.Hessels D., Schalken J.A. The use of PCA3 in the diagnosis of prostate cancer. Nat. Rev. Urol. 2009;6:255–261. doi: 10.1038/nrurol.2009.40. [DOI] [PubMed] [Google Scholar]
- 88.Hessels D., Klein Gunnewiek J.M., van Oort I., Karthaus H.F., van Leenders G.J., van Balken B., Kiemeney L.A., Witjes J.A., Schalken J.A. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur. Urol. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. ; discussion 15–16. [DOI] [PubMed] [Google Scholar]
- 89.Van Gils M.P., Hessels D., van Hooij O., Jannink S.A., Peelen W.P., Hanssen S.L., Witjes J.A., Cornel E.B., Karthaus H.F., Smits G.A., et al. The time-resolved fluorescence-based PCA3 test on urinary sediments after digital rectal examination; a Dutch multicenter validation of the diagnostic performance. Clin. Cancer Res. 2007;13:939–943. doi: 10.1158/1078-0432.CCR-06-2679. [DOI] [PubMed] [Google Scholar]
- 90.Groskopf J., Aubin S.M., Deras I.L., Blase A., Bodrug S., Clark C., Brentano S., Mathis J., Pham J., Meyer T., et al. APTIMA PCA3 molecular urine test: Development of a method to aid in the diagnosis of prostate cancer. Clin. Chem. 2006;52:1089–1095. doi: 10.1373/clinchem.2005.063289. [DOI] [PubMed] [Google Scholar]
- 91.Deras I.L., Aubin S.M., Blase A., Day J.R., Koo S., Partin A.W., Ellis W.J., Marks L.S., Fradet Y., Rittenhouse H., et al. PCA3: A molecular urine assay for predicting prostate biopsy outcome. J. Urol. 2008;179:1587–1592. doi: 10.1016/j.juro.2007.11.038. [DOI] [PubMed] [Google Scholar]
- 92.Ploussard G., Durand X., Xylinas E., Moutereau S., Radulescu C., Forgue A., Nicolaiew N., Terry S., Allory Y., Loric S., et al. Prostate cancer antigen 3 score accurately predicts tumor volume and might help in selecting prostate cancer patients for active surveillance. Eur. Urol. 2011;59:422–429. doi: 10.1016/j.eururo.2010.11.044. [DOI] [PubMed] [Google Scholar]
- 93.Van Gils M.P., Cornel E.B., Hessels D., Peelen W.P., Witjes J.A., Mulders P.F., Rittenhouse H.G., Schalken J.A. Molecular PCA3 diagnostics on prostatic fluid. Prostate. 2007;67:881–887. doi: 10.1002/pros.20564. [DOI] [PubMed] [Google Scholar]
- 94.Gu Z., Thomas G., Yamashiro J., Shintaku I.P., Dorey F., Raitano A., Witte O.N., Said J.W., Loda M., Reiter R.E. Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene. 2000;19:1288–1296. doi: 10.1038/sj.onc.1203426. [DOI] [PubMed] [Google Scholar]
- 95.Gu Z., Yamashiro J., Kono E., Reiter R.E. Anti-prostate stem cell antigen monoclonal antibody 1G8 induces cell death in vitro and inhibits tumor growth in vivo via a Fc-independent mechanism. Cancer Res. 2005;65:9495–9500. doi: 10.1158/0008-5472.CAN-05-2086. [DOI] [PubMed] [Google Scholar]
- 96.Ameri A., Alidoosti A., Hosseini S.Y., Parvin M., Emranpour M.H., Taslimi F., Salehi E., Fadavip P. Prognostic value of promoter hypermethylation of retinoic acid receptor beta (RARB) and CDKN2 (p16/MTS1) in prostate cancer. Chin. J. Cancer Res. 2011;23:306–311. doi: 10.1007/s11670-011-0306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Daniunaite K., Berezniakovas A., Jankevicius F., Laurinavicius A., Lazutka J.R., Jarmalaite S. Frequent methylation of RASSF1 and RARB in urine sediments from patients with early stage prostate cancer. Medicina (Kaunas) 2011;47:147–153. [PubMed] [Google Scholar]
- 98.Sreekumar A., Poisson L.M., Rajendiran T.M., Khan A.P., Cao Q., Yu J., Laxman B., Mehra R., Lonigro R.J., Li Y., et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 99.Tomlins S.A., Rhodes D.R., Yu J., Varambally S., Mehra R., Perner S., Demichelis F., Helgeson B.E., Laxman B., Morris D.S., et al. The role of SPINK1 in ETS rearrangement-negative prostate cancers. Cancer Cell. 2008;13:519–528. doi: 10.1016/j.ccr.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meid F.H., Gygi C.M., Leisinger H.J., Bosman F.T., Benhattar J. The use of telomerase activity for the detection of prostatic cancer cells after prostatic massage. J. Urol. 2001;165:1802–1805. [PubMed] [Google Scholar]
- 101.March-Villalba J.A., Martinez-Jabaloyas J.M., Herrero M.J., Santamaria J., Alino S.F., Dasi F. Plasma hTERT mRNA discriminates between clinically localized and locally advanced disease and is a predictor of recurrence in prostate cancer patients. Expert Opin. Biol. Ther. 2012;12:S69–S77. doi: 10.1517/14712598.2012.685716. [DOI] [PubMed] [Google Scholar]
- 102.Shariat S.F., Walz J., Roehrborn C.G., Montorsi F., Jeldres C., Saad F., Karakiewicz P.I. Early postoperative plasma transforming growth factor-beta1 is a strong predictor of biochemical progression after radical prostatectomy. J. Urol. 2008;179:1593–1597. doi: 10.1016/j.juro.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 103.Ivanovic V., Melman A., Davis-Joseph B., Valcic M., Geliebter J. Elevated plasma levels of TGF-beta 1 in patients with invasive prostate cancer. Nat. Med. 1995;1:282–284. doi: 10.1038/nm0495-282. [DOI] [PubMed] [Google Scholar]
- 104.Hessels D., Smit F.P., Verhaegh G.W., Witjes J.A., Cornel E.B., Schalken J.A. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin. Cancer Res. 2007;13:5103–5108. doi: 10.1158/1078-0432.CCR-07-0700. [DOI] [PubMed] [Google Scholar]
- 105.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 106.Mosquera J.M., Mehra R., Regan M.M., Perner S., Genega E.M., Bueti G., Shah R.B., Gaston S., Tomlins S.A., Wei J.T., et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin. Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McCabe N.P., Angwafo F.F., III, Zaher A., Selman S.H., Kouinche A., Jankun J. Expression of soluble urokinase plasminogen activator receptor may be related to outcome in prostate cancer patients. Oncol. Rep. 2000;7:879–882. doi: 10.3892/or.7.4.879. [DOI] [PubMed] [Google Scholar]
- 108.Shariat S.F., Roehrborn C.G., McConnell J.D., Park S., Alam N., Wheeler T.M., Slawin K.M. Association of the circulating levels of the urokinase system of plasminogen activation with the presence of prostate cancer and invasion, progression, and metastasis. J. Clin. Oncol. 2007;25:349–355. doi: 10.1200/JCO.2006.05.6853. [DOI] [PubMed] [Google Scholar]
- 109.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 110.Roupret M., Hupertan V., Yates D.R., Catto J.W., Rehman I., Meuth M., Ricci S., Lacave R., Cancel-Tassin G., de la Taille A., et al. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin. Cancer Res. 2007;13:1720–1725. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]
- 111.Nakayama M., Gonzalgo M.L., Yegnasubramanian S., Lin X., de Marzo A.M., Nelson W.G. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J. Cell Biochem. 2004;91:540–552. doi: 10.1002/jcb.10740. [DOI] [PubMed] [Google Scholar]
- 112.Hoque M.O., Topaloglu O., Begum S., Henrique R., Rosenbaum E., van Criekinge W., Westra W.H., Sidransky D. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J. Clin. Oncol. 2005;23:6569–6575. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 113.Costa V.L., Henrique R., Danielsen S.A., Eknaes M., Patricio P., Morais A., Oliveira J., Lothe R.A., Teixeira M.R., Lind G.E., et al. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics. 2011;6:1120–1130. doi: 10.4161/epi.6.9.16376. [DOI] [PubMed] [Google Scholar]
- 114.Beckett M.L., Cazares L.H., Vlahou A., Schellhammer P.F., Wright G.L., Jr Prostate-specific membrane antigen levels in sera from healthy men and patients with benign prostate hyperplasia or prostate cancer. Clin. Cancer Res. 1999;5:4034–4040. [PubMed] [Google Scholar]
- 115.Sokoloff R.L., Norton K.C., Gasior C.L., Marker K.M., Grauer L.S. A dual-monoclonal sandwich assay for prostate-specific membrane antigen: Levels in tissues, seminal fluid and urine. Prostate. 2000;43:150–157. doi: 10.1002/(sici)1097-0045(20000501)43:2<150::aid-pros10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 116.Xiao Z., Adam B.L., Cazares L.H., Clements M.A., Davis J.W., Schellhammer P.F., Dalmasso E.A., Wright G.L., Jr Quantitation of serum prostate-specific membrane antigen by a novel protein biochip immunoassay discriminates benign from malignant prostate disease. Cancer Res. 2001;61:6029–6033. [PubMed] [Google Scholar]
- 117.Talesa V.N., Antognelli C., del Buono C., Stracci F., Serva M.R., Cottini E., Mearini E. Diagnostic potential in prostate cancer of a panel of urinary molecular tumor markers. Cancer Biomark. 2009;5:241–251. doi: 10.3233/CBM-2009-0109. [DOI] [PubMed] [Google Scholar]
- 118.Mitchell P.J., Welton J., Staffurth J., Court J., Mason M.D., Tabi Z., Clayton A. Can urinary exosomes act as treatment response markers in prostate cancer? J. Transl. Med. 2009;7:4. doi: 10.1186/1479-5876-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rigau M., Ortega I., Mir M.C., Ballesteros C., Garcia M., Llaurado M., Colas E., Pedrola N., Montes M., Sequeiros T., et al. A Three-Gene panel on urine increases PSA specificity in the detection of prostate cancer. Prostate. 2011;71:1736–1745. doi: 10.1002/pros.21390. [DOI] [PubMed] [Google Scholar]
- 120.Magi-Galluzzi C., Tsusuki T., Elson P., Simmerman K., LaFargue C., Esgueva R., Klein E., Rubin M.A., Zhou M. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71:489–497. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 121.Rostad K., Hellwinkel O.J., Haukaas S.A., Halvorsen O.J., Oyan A.M., Haese A., Budaus L., Albrecht H., Akslen L.A., Schlomm T., et al. TMPRSS2:ERG fusion transcripts in urine from prostate cancer patients correlate with a less favorable prognosis. APMIS. 2009;117:575–582. doi: 10.1111/j.1600-0463.2009.02517.x. [DOI] [PubMed] [Google Scholar]
- 122.Demichelis F., Fall K., Perner S., Andren O., Schmidt F., Setlur S.R., Hoshida Y., Mosquera J.M., Pawitan Y., Lee C., et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–4599. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 123.Bader A.G., Brown D., Stoudemire J., Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Catto J.W., Miah S., Owen H.C., Bryant H., Myers K., Dudziec E., Larre S., Milo M., Rehman I., Rosario D.J., et al. Distinct microRNA alterations characterize high- and low-grade bladder cancer. Cancer Res. 2009;69:8472–8481. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell. Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 127.Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell. Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 128.Gangaraju V.K., Lin H. MicroRNAs: Key regulators of stem cells. Nat. Rev. Mol. Cell. Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuner R., Brase J.C., Sultmann H., Wuttig D. microRNA biomarkers in body fluids of prostate cancer patients. Methods. 2013;59:132–137. doi: 10.1016/j.ymeth.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 131.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 132.Gordanpour A., Nam R.K., Sugar L., Seth A. MicroRNAs in prostate cancer: From biomarkers to molecularly-based therapeutics. Prostate Cancer Prostatic Dis. 2012;15:314–319. doi: 10.1038/pcan.2012.3. [DOI] [PubMed] [Google Scholar]
- 133.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O’Briant K.C., Allen A., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Brase J.C., Johannes M., Schlomm T., Falth M., Haese A., Steuber T., Beissbarth T., Kuner R., Sultmann H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer. 2011;128:608–616. doi: 10.1002/ijc.25376. [DOI] [PubMed] [Google Scholar]
- 136.Yamada Y., Enokida H., Kojima S., Kawakami K., Chiyomaru T., Tatarano S., Yoshino H., Kawahara K., Nishiyama K., Seki N., et al. MiR-96 and miR-183 detection in urine serve as potential tumor markers of urothelial carcinoma: Correlation with stage and grade, and comparison with urinary cytology. Cancer Sci. 2011;102:522–529. doi: 10.1111/j.1349-7006.2010.01816.x. [DOI] [PubMed] [Google Scholar]
- 137.Bryant R.J., Pawlowski T., Catto J.W., Marsden G., Vessella R.L., Rhees B., Kuslich C., Visakorpi T., Hamdy F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer. 2012;106:768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sapre N., Selth L.A. Circulating MicroRNAs as biomarkers of prostate cancer: The state of play. Prostate Cancer. 2013;2013:539680. doi: 10.1155/2013/539680. :1–539680:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Costa F.F. Non-coding RNAs: New players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 140.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 141.Szell M., Bata-Csorgo Z., Kemeny L. The enigmatic world of mRNA-like ncRNAs: Their role in human evolution and in human diseases. Semin. Cancer Biol. 2008;18:141–148. doi: 10.1016/j.semcancer.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 142.Turner A.M., Morris K.V. Controlling transcription with noncoding RNAs in mammalian cells. Biotechniques. 2010;48:ix–xvi. doi: 10.2144/000113442. [DOI] [PubMed] [Google Scholar]
- 143.Pavlou M.P., Diamandis E.P. The cancer cell secretome: A good source for discovering biomarkers? J. Proteomics. 2010;73:1896–1906. doi: 10.1016/j.jprot.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 144.Makridakis M., Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J. Proteomics. 2010;73:2291–2305. doi: 10.1016/j.jprot.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 145.Xue H., Lu B., Zhang J., Wu M., Huang Q., Wu Q., Sheng H., Wu D., Hu J., Lai M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J. Proteome Res. 2009;9:545–555. doi: 10.1021/pr9008817. [DOI] [PubMed] [Google Scholar]
- 146.Bolduc S., Lacombe L., Naud A., Gregoire M., Fradet Y., Tremblay R.R. Urinary PSA: A potential useful marker when serum PSA is between 2.5 ng/mL and 10 ng/mL. Can. Urol. Assoc. J. 2007;1:377–381. doi: 10.5489/cuaj.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Takayama T.K., Vessella R.L., Brawer M.K., True L.D., Noteboom J., Lange P.H. Urinary prostate specific antigen levels after radical prostatectomy. J. Urol. 1994;151:82–87. doi: 10.1016/s0022-5347(17)34877-2. [DOI] [PubMed] [Google Scholar]
- 148.DeVere White R.W., Meyers F.J., Soares S.E., Miller D.G., Soriano T.F. Urinary prostate specific antigen levels: Role in monitoring the response of prostate cancer to therapy. J. Urol. 1992;147:947–951. doi: 10.1016/s0022-5347(17)37430-x. [DOI] [PubMed] [Google Scholar]
- 149.Pannek J., Rittenhouse H.G., Evans C.L., Finlay J.A., Bruzek D.J., Cox J.L., Chan D.W., Subong E.N., Partin A.W. Molecular forms of prostate-specific antigen and human kallikrein 2 (hK2) in urine are not clinically useful for early detection and staging of prostate cancer. Urology. 1997;50:715–721. doi: 10.1016/S0090-4295(97)00324-5. [DOI] [PubMed] [Google Scholar]
- 150.Malavaud B., Salama G., Miedouge M., Vincent C., Rischmann P., Sarramon J.P., Serre G. Influence of digital rectal massage on urinary prostate-specific antigen: Interest for the detection of local recurrence after radical prostatectomy. Prostate. 1998;34:23–28. doi: 10.1002/(sici)1097-0045(19980101)34:1<23::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 151.Schostak M., Schwall G.P., Poznanovic S., Groebe K., Muller M., Messinger D., Miller K., Krause H., Pelzer A., Horninger W., et al. Annexin A3 in urine: A highly specific noninvasive marker for prostate cancer early detection. J. Urol. 2009;181:343–353. doi: 10.1016/j.juro.2008.08.119. [DOI] [PubMed] [Google Scholar]
- 152.Kulasingam V., Diamandis E.P. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008;5:588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 153.Decramer S., Gonzalez de Peredo A., Breuil B., Mischak H., Monsarrat B., Bascands J.L., Schanstra J.P. Urine in clinical proteomics. Mol. Cell Proteomics. 2008;7:1850–1862. doi: 10.1074/mcp.R800001-MCP200. [DOI] [PubMed] [Google Scholar]
- 154.Rehman I., Azzouzi A.R., Catto J.W., Allen S., Cross S.S., Feeley K., Meuth M., Hamdy F.C. Proteomic analysis of voided urine after prostatic massage from patients with prostate cancer: A pilot study. Urology. 2004;64:1238–1243. doi: 10.1016/j.urology.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 155.Theodorescu D., Fliser D., Wittke S., Mischak H., Krebs R., Walden M., Ross M., Eltze E., Bettendorf O., Wulfing C., et al. Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis. 2005;26:2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- 156.Theodorescu D., Schiffer E., Bauer H.W., Douwes F., Eichhorn F., Polley R., Schmidt T., Schofer W., Zurbig P., Good D.M., et al. Discovery and validation of urinary biomarkers for prostate cancer. Proteomics Clin. Appl. 2008;2:556–570. doi: 10.1002/prca.200780082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.M’Koma A.E., Blum D.L., Norris J.L., Koyama T., Billheimer D., Motley S., Ghiassi M., Ferdowsi N., Bhowmick I., Chang S.S., et al. Detection of pre-neoplastic and neoplastic prostate disease by MALDI profiling of urine. Biochem. Biophys. Res. Commun. 2007;353:829–834. doi: 10.1016/j.bbrc.2006.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Okamoto A., Yamamoto H., Imai A., Hatakeyama S., Iwabuchi I., Yoneyama T., Hashimoto Y., Koie T., Kamimura N., Mori K., et al. Protein profiling of post-prostatic massage urine specimens by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry to discriminate between prostate cancer and benign lesions. Oncol. Rep. 2009;21:73–79. [PubMed] [Google Scholar]
- 159.Gamagedara S., Kaczmarek A.T., Jiang Y., Cheng X., Rupasinghe M., Ma Y. Validation study of urinary metabolites as potential biomarkers for prostate cancer detection. Bioanalysis. 2012;4:1175–1183. doi: 10.4155/bio.12.92. [DOI] [PubMed] [Google Scholar]
- 160.Jentzmik F., Stephan C., Miller K., Schrader M., Erbersdobler A., Kristiansen G., Lein M., Jung K. Sarcosine in urine after digital rectal examination fails as a marker in prostate cancer detection and identification of aggressive tumors. Eur. Urol. 2010;58:12–18. doi: 10.1016/j.eururo.2010.01.035. ; discussion 20–21. [DOI] [PubMed] [Google Scholar]
- 161.Landers K.A., Burger M.J., Tebay M.A., Purdie D.M., Scells B., Samaratunga H., Lavin M.F., Gardiner R.A. Use of multiple biomarkers for a molecular diagnosis of prostate cancer. Int. J. Cancer. 2005;114:950–956. doi: 10.1002/ijc.20760. [DOI] [PubMed] [Google Scholar]
- 162.Schmidt U., Fuessel S., Koch R., Baretton G.B., Lohse A., Tomasetti S., Unversucht S., Froehner M., Wirth M.P., Meye A. Quantitative multi-gene expression profiling of primary prostate cancer. Prostate. 2006;66:1521–1534. doi: 10.1002/pros.20490. [DOI] [PubMed] [Google Scholar]
- 163.Etzioni R., Kooperberg C., Pepe M., Smith R., Gann P.H. Combining biomarkers to detect disease with application to prostate cancer. Biostatistics. 2003;4:523–538. doi: 10.1093/biostatistics/4.4.523. [DOI] [PubMed] [Google Scholar]
- 164.Baden J., Green G., Painter J., Curtin K., Markiewicz J., Jones J., Astacio T., Canning S., Quijano J., Guinto W., et al. Multicenter evaluation of an investigational prostate cancer methylation assay. J. Urol. 2009;182:1186–1193. doi: 10.1016/j.juro.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 165.Vener T., Derecho C., Baden J., Wang H., Rajpurohit Y., Skelton J., Mehrotra J., Varde S., Chowdary D., Stallings W., et al. Development of a multiplexed urine assay for prostate cancer diagnosis. Clin. Chem. 2008;54:874–882. doi: 10.1373/clinchem.2007.094912. [DOI] [PubMed] [Google Scholar]
- 166.Payne S.R., Serth J., Schostak M., Kamradt J., Strauss A., Thelen P., Model F., Day J.K., Liebenberg V., Morotti A., et al. DNA methylation biomarkers of prostate cancer: Confirmation of candidates and evidence urine is the most sensitive body fluid for non-invasive detection. Prostate. 2009;69:1257–1269. doi: 10.1002/pros.20967. [DOI] [PubMed] [Google Scholar]
- 167.Ouyang B., Bracken B., Burke B., Chung E., Liang J., Ho S.M. A duplex quantitative polymerase chain reaction assay based on quantification of alpha-methylacyl-CoA racemase transcripts and prostate cancer antigen 3 in urine sediments improved diagnostic accuracy for prostate cancer. J. Urol. 2009;181:2508–2513. doi: 10.1016/j.juro.2009.01.110. ; discussion 2513–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Salami S.S., Schmidt F., Laxman B., Regan M.M., Rickman D.S., Scherr D., Bueti G., Siddiqui J., Tomlins S.A., Wei J.T., et al. Combining urinary detection of TMPRSS2:ERG and PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol. Oncol. 2011 doi: 10.1016/j.urolonc.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jamaspishvili T., Kral M., Khomeriki I., Student V., Kolar Z., Bouchal J. Urine markers in monitoring for prostate cancer. Prostate Cancer Prostatic Dis. 2011;13:12–19. doi: 10.1038/pcan.2009.31. [DOI] [PubMed] [Google Scholar]
- 170.Nguyen P.N., Violette P., Chan S., Tanguay S., Kassouf W., Aprikian A., Chen J.Z. A panel of TMPRSS2:ERG fusion transcript markers for urine-based prostate cancer detection with high specificity and sensitivity. Eur. Urol. 2011;59:407–414. doi: 10.1016/j.eururo.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 171.Tomlins S.A., Aubin S.M., Siddiqui J., Lonigro R.J., Sefton-Miller L., Miick S., Williamsen S., Hodge P., Meinke J., Blase A., et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med. 2011;3:94r. doi: 10.1126/scitranslmed.3001970. a72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Cao D.L., Ye D.W., Zhang H.L., Zhu Y., Wang Y.X., Yao X.D. A multiplex model of combining gene-based, protein-based, and metabolite-based with positive and negative markers in urine for the early diagnosis of prostate cancer. Prostate. 2011;71:700–710. doi: 10.1002/pros.21286. [DOI] [PubMed] [Google Scholar]
- 173.Prior C., Guillen-Grima F., Robles J.E., Rosell D., Fernandez-Montero J.M., Agirre X., Catena R., Calvo A. Use of a combination of biomarkers in serum and urine to improve detection of prostate cancer. World J. Urol. 2010;2(8):681–686. doi: 10.1007/s00345-010-0583-x. [DOI] [PubMed] [Google Scholar]
- 174.Thery C., Zitvogel L., Amigorena S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 175.Simpson R.J., Jensen S.S., Lim J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 176.Guescini M., Genedani S., Stocchi V., Agnati L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- 177.Keller S., Ridinger J., Rupp A.K., Janssen J.W., Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011;9:86. doi: 10.1186/1479-5876-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhou H., Yuen P.S., Pisitkun T., Gonzales P.A., Yasuda H., Dear J.W., Gross P., Knepper M.A., Star R.A. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nilsson J., Skog J., Nordstrand A., Baranov V., Mincheva-Nilsson L., Breakefield X.O., Widmark A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hessvik N.P., Sandvig K., Llorente A. Exosomal miRNAs as biomarkers for prostate cancer. Front Genet. 2013;4:36. doi: 10.3389/fgene.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Laxman B., Tomlins S.A., Mehra R., Morris D.S., Wang L., Helgeson B.E., Shah R.B., Rubin M.A., Wei J.T., Chinnaiyan A.M. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8:885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Makarov D.V., Loeb S., Getzenberg R.H., Partin A.W. Biomarkers for prostate cancer. Annu. Rev. Med. 2009;60:139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 183.Ploussard G., de la Taille A. Urine biomarkers in prostate cancer. Nat. Rev. Urol. 2010;7:101–109. doi: 10.1038/nrurol.2009.261. [DOI] [PubMed] [Google Scholar]