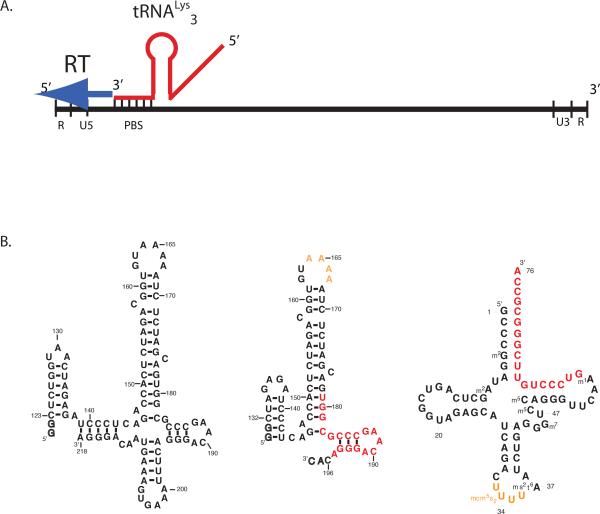
Figure 1.
(A.) Schematic of the reverse transcription initiation complex within the HIV genome. The RNA genome of HIV RNA has the standard structure for retroviral genomes, with long terminal repeat regions (U5, U3, R), and the primer binding site (PBS) near the 5'end. Host tRNALys3 interacts with the PBS through an 18 base-pair interaction, providing the 3'-OH group to initiate reverse transcription by viral reverse transcriptase (RT).
(B.) RNA oligonucleotides corresponding to HIV-1 genomic RNAs used in the current study (left) a 99 nt and (right) 69 nt RNA with numbering according to the Mal isolate. Additional nucleotides added at the 5'end for transcription with T7 RNA polymerase are indicated as outlines. The primer binding site region of 18bp of complementarity with tRNA is highlighted in red, whereas the A-rich loop is in yellow. (c) Secondary structure of human tRNALys3 with modified nucleotides; Coloring as in (b).