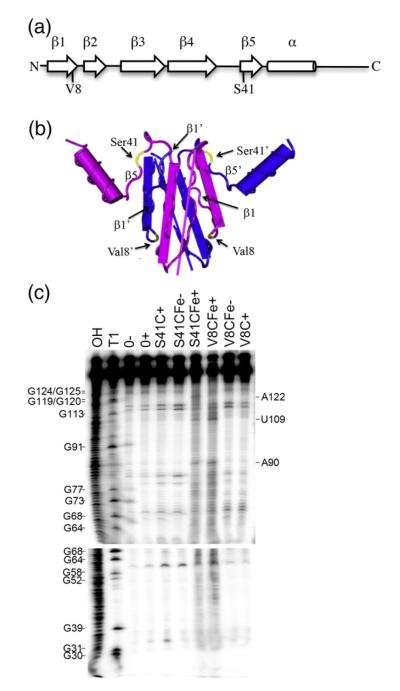
Fig. 3.
Footprinting of the moaA mRNA leader RNA with CsrA-FeBABE. (a) Cysteine residues were singly introduced at two different positions within the CsrA protein, valine 8 (V8C) or serine 41 (S41C), and derivatized with the FeBABE reagent. (b) The two polypeptides of the CsrA dimer are shown as blue or purple. Note that the β1 and β5 strands of opposite polypeptides of the dimer lie parallel with each other in the three-dimensional structure of the protein.9 Residues Val8 and Ser41 are shown in yellow. (c) Footprinting of moaA leader RNA. 32P-labeled moaA RNA was incubated with no CsrA (0), CsrA S41C (500 nM) or CsrA V8C (250 nM) in the absence (−) or presence (+) of cleaving reagents, ascorbic acid and hydrogen peroxide. OH and T1 indicate moaA RNA alkaline hydrolysis and partial RNase T1 ladders, respectively. Positions of strand scission are indicated on the right. Numbering is from the start of the moaA transcript shown in Fig. 1a.