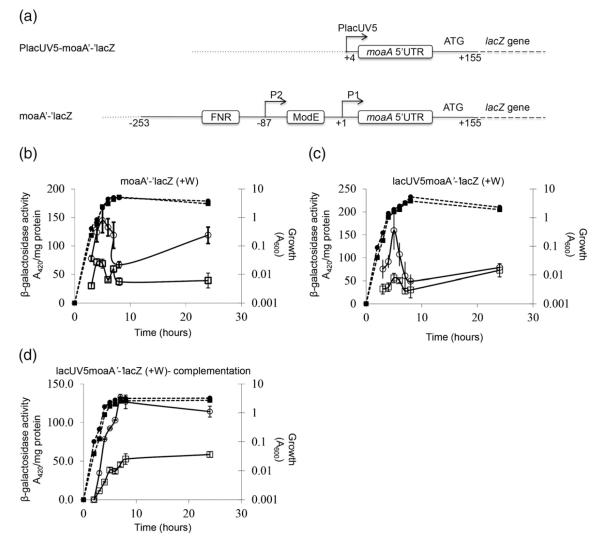
Fig. 5.
Effects of CsrA on expression of chromosomal moaA‘-’lacZ and PlacUV5-moaA‘-’lacZ fusions. (a) The PlacUV5-moaA‘-’lacZ posttranscriptional fusion and the moaA‘-’lacZ translational fusion are depicted. Binding sites for FNR and ModE transcription factors in the latter fusion are marked. Vector DNA (…) and the junctions between moaA DNA (—) and lacZ DNA (—) are also indicated. (b–d) For expression studies, cultures were harvested at various time points after inoculation (T=0) and assayed for β-galactosidase specific activity (A420 per milligram of protein). Isogenic strains were from the CF7789 strain background. We added 10 mM sodium tungstate to derepress Moco-mediated regulation. Values represent independent experiments performed in triplicate, and error bars depict standard deviation of the mean. Growth (broken lines) and β-galactosidase-specific activity (continuous lines) are presented. (b) Activity from a moaA‘-’lacZ translational fusion. CF7789, (●); csrA, (∎). (c) Activity from a PlacUV5-moaA‘-’lacZ posttranscriptional fusion. CF7789, (●); csrA, (∎). (d) Complementation analysis of expression from a PlacUV5moaA‘-’lacZ fusion in a csrA::kan strain containing pBR322 (empty vector) (∎) or the csrA-bearing plasmid pCRA16 (●).