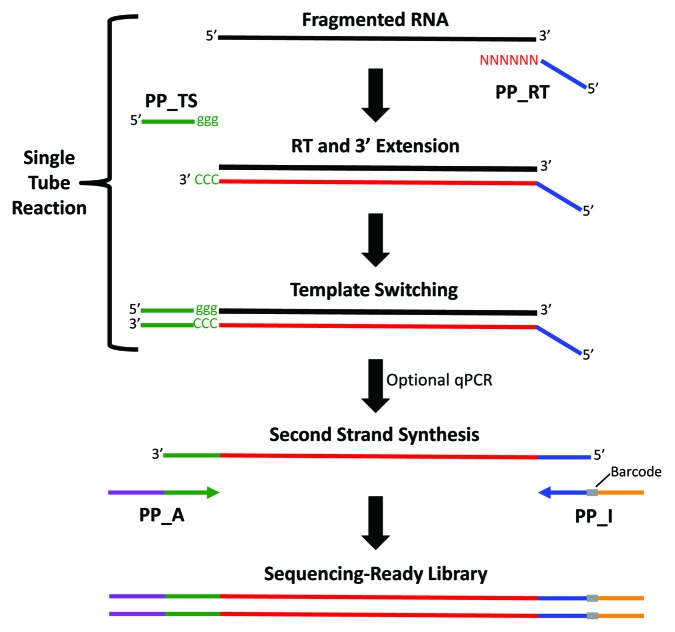
Figure 1. Overview of the Peregrine RNA-Seq library preparation technique. Chemically fragmented RNA anneals with the random hexamer end of the primer PP_RT, and MMLV reverse transcriptase polymerizes the first cDNA strand. Upon reaching the end of the RNA template, the enzyme adds the untemplated sequence CCC to the 3′ end of the new cDNA strand. This serves as the annealing site for the complementary ribonucleotide sequence (ggg) of oligonucleotide PP_TS. At this point the enzyme switches templates, extending the first cDNA strand to incorporate sequence complementary to the PP_TS template. A qPCR assay can be performed to determine the number of PCR cycles required for optimal second strand synthesis and library amplification. During second strand synthesis, Illumina sequencing adaptors (one including a barcode) are incorporated at the ends of the cDNA. Depletion of rRNA-derived sequences may be performed prior to RNA fragmentation (Ribo-Zero) or following PCR (DSN-mediated normalization).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.