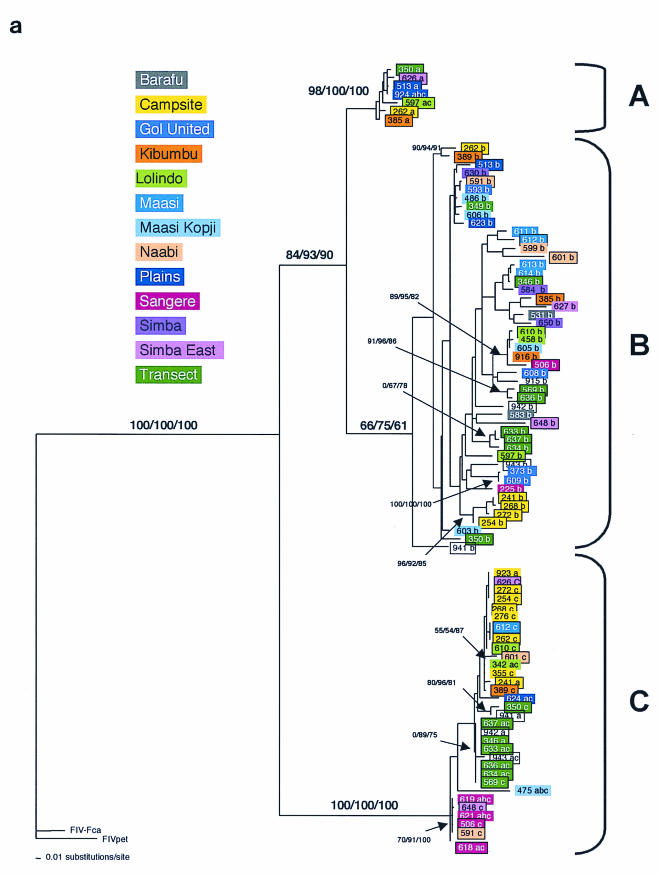
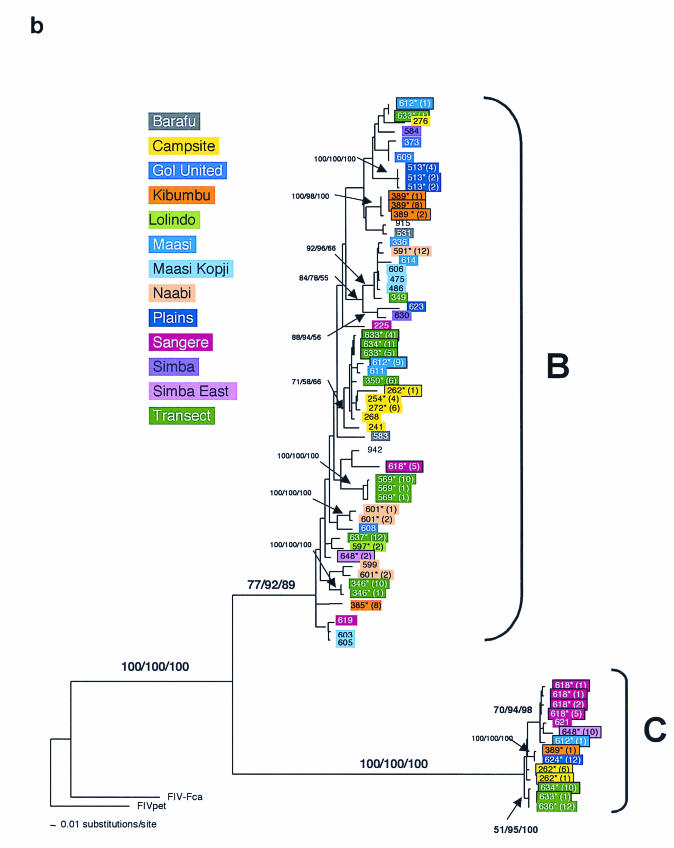
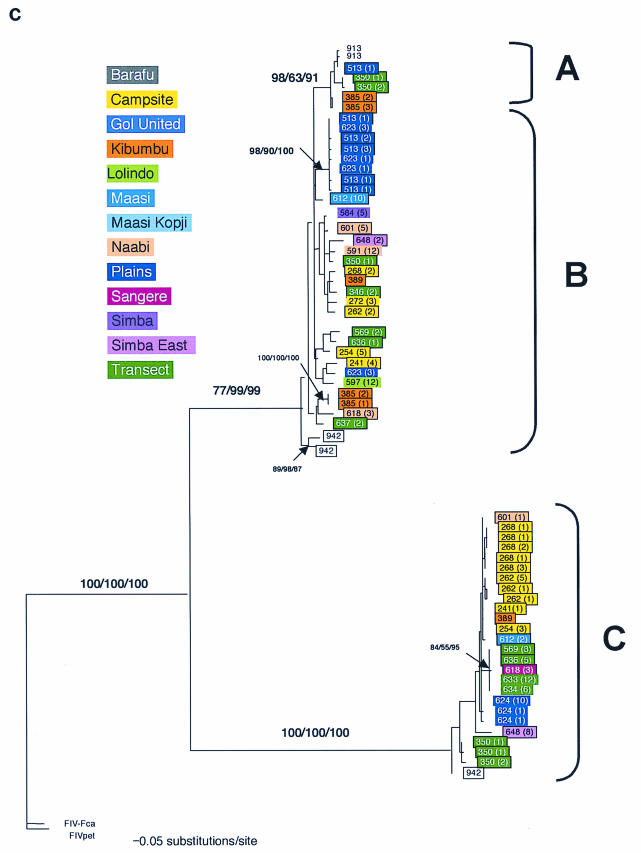
FIG.3.
Phylogenetic tree for FIV-Ple nucleotide sequences. Shown here in each case is the single maximum likelihood (ML) tree. Branch lengths represent percent sequence divergence based on the model of sequence evolution specified by ModelTest (39). Minimum evolution estimated by neighbor-joining (NJ) and maximum parsimony (MP) trees gave similar topologies, and bootstrap values are included at all nodes with bootstrap support of >70 (ML/NJ/MP). Taxa are designated with the source lion identification number. Boxed taxa indicate individuals with FIV sequences that fall into more than one clade. Cloned sequences are followed by an asterisk, and the number of clones with that sequence is given in parentheses. Pride affiliations are designated by color. (a) pol-RT, 337 bp. Maximum likelihood tree under the HKY+I+G model of sequence evolution with an estimated shape parameter of 0.6272, an estimated transition/transversion ratio of 4.0911, and an estimated proportion of invariant sites of 0.3750 (−ln likelihood = 3,978.9614). Taxa are followed by a lowercase letter indicating the internal subtype-specific primer used (a, b, or c) (Fig. 1A). In most instances, an RT-subtype “a” primer set produced an RT-subtype A sequence, RT-subtype “b” primers produced an RT-subtype B sequence, and “c” primers produced a C sequence. Reference sequences for the three previously identified FIV-Ple subtypes are as follows: A (Ple-241 and Ple-385), B (Ple-254 and Ple-272), and C (Ple-355 and Ple-389). (b) pol-RNase, 730 bp. The maximum likelihood tree under the GTR+I+G model of sequence evolution has an estimated shape parameter of 0.4274 and an estimated substitution matrix as follows: A/C = 2.0073, A/G = 7.902, A/T = 1.4491, C/G = 2.6080, C/T = 14.7031, and an estimated proportion of invariant sites of 0.1153 (−ln likelihood = 6,951.6733). (c) gag cloned sequences, 444 bp. Boxed taxa with no numbers in parentheses are the result of different sequences obtained at different temperatures in a gradient. The maximum likelihood tree under the GTR+G model of sequence evolution has an estimated shape parameter of 0.3584 and an estimated substitution matrix as follows: A/C = 2.3398, A/G = 9.8759, A/T = 2.6700, C/G = 2.8285, C/T = 16.5121 (−ln likelihood = 3,582.6128). Deletion of 13 amino acid residues in FIV-Ple C is excluded in this analysis (see text).