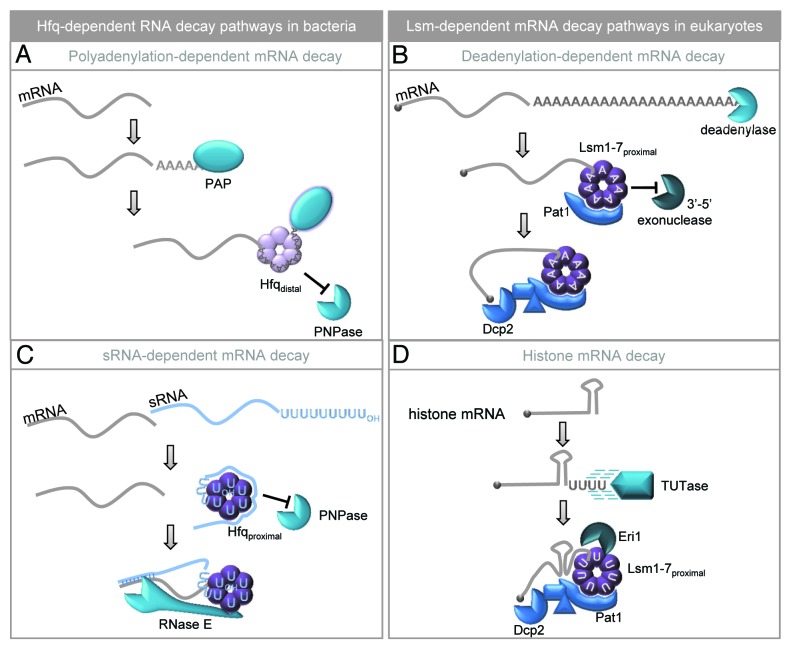
Figure 2. Hfq and Lsm proteins bind single-stranded 3′ ends and activate RNA decay. (A) Poly(A) polymerase (PAP) acts distributively to add adenosine residues until the poly(A) tail reaches sufficient length to recruit Hfq. Hfq binds poly(A) through its distal surface and stimulates PAP activity to extend the poly(A) tail. This interaction also prevents the 3′-5′ exonuclease, polynucleotide phosphorylase (PNPase), from attacking the 3′ end of the transcript.35-37 (B) In eukaryotic cells, poly(A) tails are shortened by deadenylase enzymes prior to decay.38 Once the tail is short enough (~10–12 residues), Lsm1-7 associates using the proximal binding surface and Pat1 is also recruited.78 The Lsm-Pat1 complex prevents further shortening from the 3′ end while activating decapping at the 5′ end by interacting with a number of accessory factors to engage the decapping enzyme (Dcp2).40 (C) Bacterial sRNAs terminate in a tract of seven to nine uridines generated through Rho-independent termination. This 3′ U-tract is recognized by Hfq through its proximal binding surface resulting in protection of the 3′OH group from attack by PNPase. Additional regions of the sRNA can bind the lateral surface of Hfq while the distal surface recognizes mRNA targets. Once the mRNA and sRNA anneal, RNase E joins the complex and cleaves both the mRNA and sRNA to initiate their degradation.42,44,45 (D) Vertebrate histone mRNAs are degraded through a pathway that requires 3′ oligouridylation by a terminal uridyltransferase (TUTase).57,58 This provides a platform to recruit the Lsm1–7/Pat1 complex which, in turn, initiates mRNA decapping.57,59 Oligouridylation of histone mRNAs also promotes 3′-5′ exonucleolytic decay as Lsm1-7 recruits the Eri1 exonuclease.60

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.