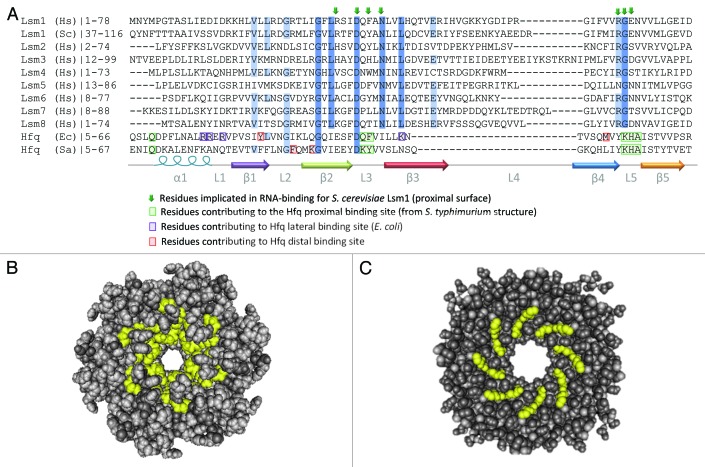
Figure 4. Conservation of RNA-binding residues in Lsm and Hfq proteins. (A) Alignment of primary sequence of Sm domains for human Lsm1-8 proteins, S. cerevisiae Lsm1 and Hfq from E. coli and S. aureus. C-terminal domains and some N-terminal sequences are excluded from this alignment for clarity. The alignment was generated using CLUSTAL-W100 and edited in JalView.101 Secondary structure is indicated below the alignment and colored as in Figure 1. Residues shown to be important for RNA-binding are highlighted (Hfq) or indicated by arrows (Lsm proteins). Conserved residues are indicated by blue shading with darker blue denoting more conservation. The accession numbers for the protein sequences used in the alignment are: H.s. Lsm1 NP_055277.1, S.c. Lsm1 NP_012411.1, H.s. Lsm2 NP_067000.1, H.s. Lsm3 NP_055278.1, H.s. Lsm4 NP_036453.1, H.s. Lsm5 NP_036454.1, H.s. Lsm6 NP_009011.1, H.s. Lsm7 NP_057283.1, H.s. Lsm8 NP_057284.1, E.c.Hfq ACE63256.1, S.a.Hfq AEW65270.1. (B) Space-filling structure of proximal surface of S. Hfq (PDB ID:2YLB). Residues involved in RNA-binding are highlighted in yellow. (C) Space-filling structure of proximal surface of an S. cerevisiae Lsm3 octamer (PDB ID: 3BW1). Residues likely to be involved in RNA recognition are highlighted in yellow. The images in (B and C) were generated using Cn3D Structure Viewer Ver 4.3.102

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.