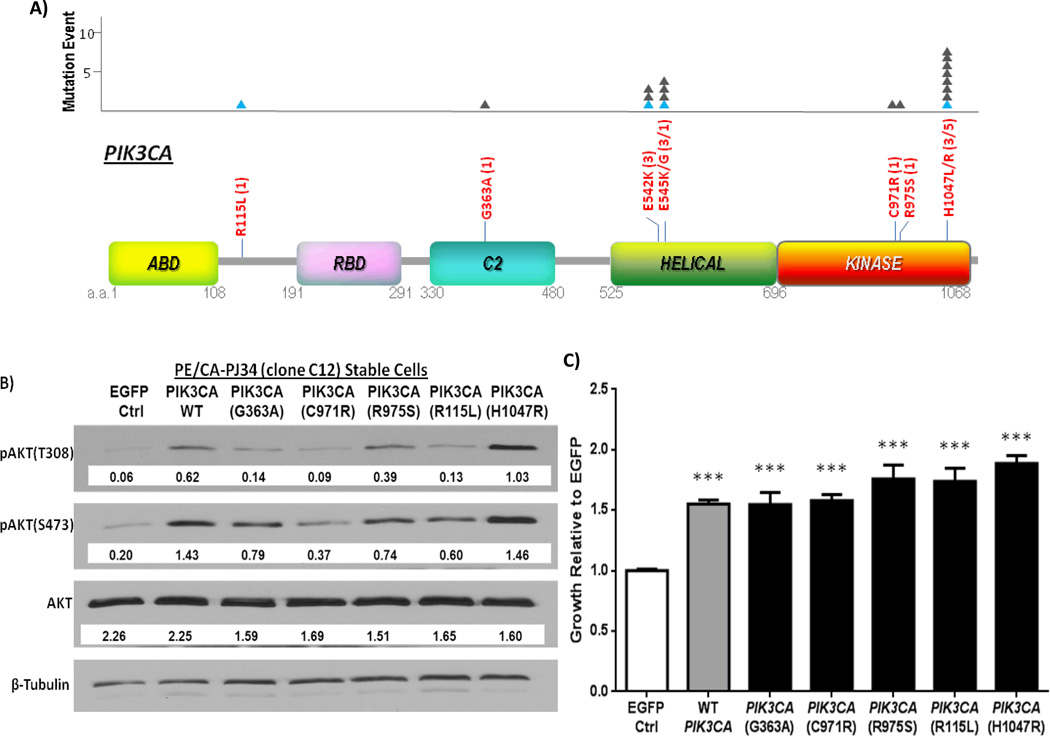
Figure 2.
PIK3CA mutations in HNSCC tumors. A) Schematic of all PIK3CA mutations found in 151 HNSCC tumors by whole exome sequencing. The amino acid (a.a.) positions of each domain is shown in grey below each domain. The number of mutational events at each site is indicated by a filled triangle (▲) in the graph above. Blue triangles indicate mutations found in HPV/HNSCC tumors. Keys: ABD: p85 binding domain; RBD: Ras binding domain; C2 Superfamily; Helical: PIK domain; Kinase: Kinase domain of PIK3CA. (B) Effects of PIK3CA mutations on PI3K signaling in HNSCC cells. WT PIK3CA, hotspot mutant H1047R, and novel mutants: R115L, G363A, C971R, and R975S were stably expressed in an HNSCC cell line harboring no endogenous mutations in the PI3K pathway, PE/CA-PJ34 (clone C12) cells, by retroviral infection. Shown here is a representative Western blot with densitometry values normalized to beta-tubulin loading controls for each engineered cell line. Increased phosphorylation of AKT at the T308 and/or S473 residue was generally observed in HNSCC cells stably expressing WT or mutant PIK3CA constructs relative to the EGFP expressing HNSCC cells, indicating enhanced activation of the PI3K signaling pathway. (C) Effects of PIK3CA mutations on HNSCC cell growth. HNSCC cells stably expressing WT or mutant PIK3CA constructs demonstrated enhanced growth at 72 hours in media with 2% FBS by MTT assay compared to cells expressing EGFP vector control (p<0.0001***). PIK3CA(H1047R) expressing cells further demonstrated enhanced growth when compared to simulated WT PIK3CA amplification (p = 0.001). Data shown here represents growth studies from three sets of independent replicate cell lines (separate infections, n=18 for each group).