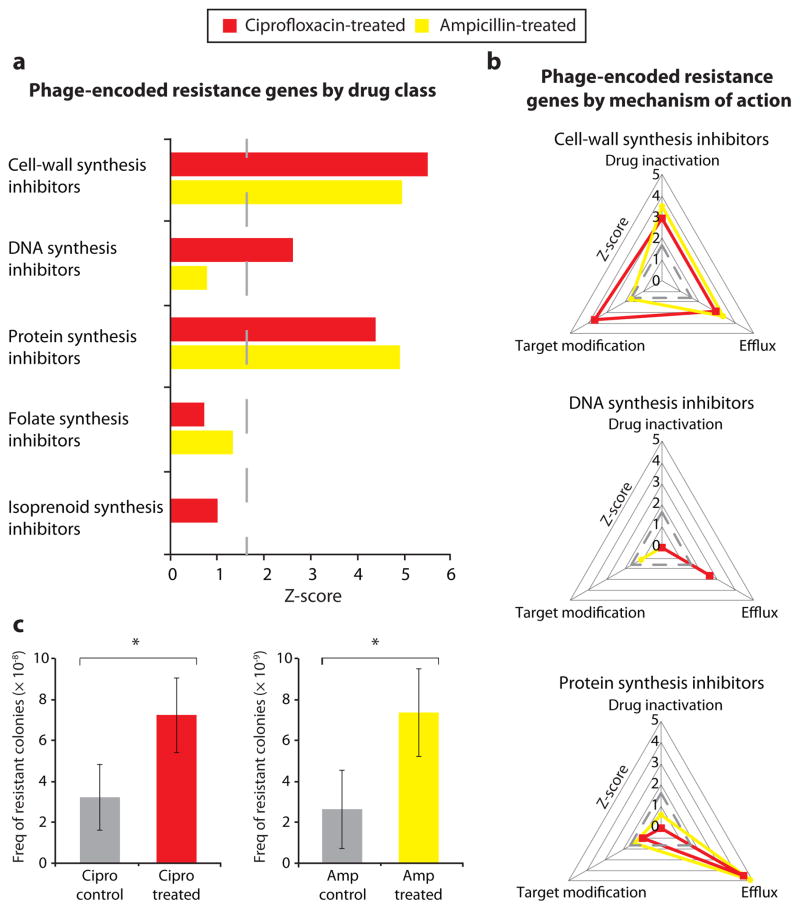
Figure 1. Antibiotic resistance is enriched in phage metagenomes following drug perturbation in mice.
a, b, Z-scores are shown for sequencing reads annotated as antibiotic resistance genes in phage from ciprofloxacin-treated mice (red) and phage from ampicillin-treated mice (yellow) in comparison to respective control mice. Dashed lines correspond to a Z-score of 1.65 (P = 0.05). Phage-encoded resistance genes were classified according to the drug class to which they confer resistance (a) and by their mechanism of resistance (b). c, Frequency of colonies resistant to ciprofloxacin (1 μg/ml) upon infection of microbiota with phage from ciprofloxacin-treated mice or phage from control mice (left), and frequency of colonies resistant to ampicillin (4 μg/ml) upon infection of microbiota with phage from ampicillin-treated mice or phage from control mice (right). P-values from Mann-Whitney U-test; n > 12. Mean ± s.e.m. *P < 0.05.