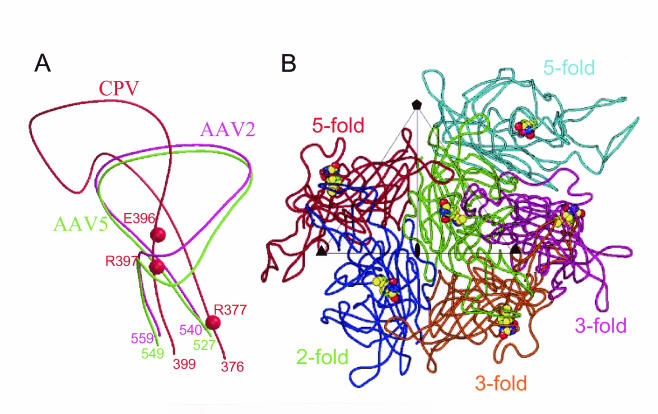
FIG. 7.
Predicted sialic acid binding residues in AAV5. (A) Close-up view of corresponding surface loops in the AAV2 (magenta), AAV5 (green), and CPV (red) capsid subunits. Sialic acid binding residues in CPV are depicted as red spheres. Additional numbered residues (magenta, green, and red) represent the N and C termini of the depicted portions of the loops for each virus. (B) View towards viral capsid surface showing a VP3 reference monomer (green) plus one twofold (blue), two threefold (magenta and orange), and two fivefold (red and cyan) VP3 molecules. Residues were deduced by superimposing the AAV2-based model of AAV5 onto the CPV structure. Predicted sialic acid binding residues (528, 546, and 547) equivalent to those involved in CPV sialic acid binding are highlighted as space-filling structures.