FIGURE 5. Functional interaction of TRF2 with RECQL4 on telomeric D-Loops containing thymine glycol.
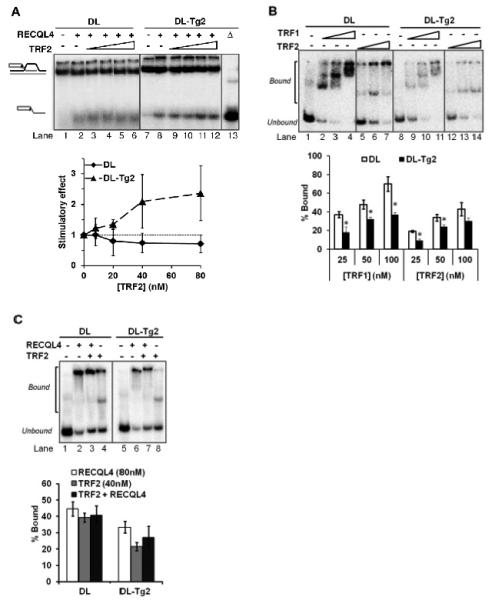
(A) Gel and quantitative analysis showing the effect of 8, 20, 40 and 80 nM TRF2 on 80 nM RECQL4 on an undamaged D-Loop, DL (lanes 1-6), and a Tg-containing D-Loop, DL-Tg2 (lanes 7-12). Δ (lane 13) indicates heat-denatured substrate. The graph represents the stimulatory effect on RECQL4’s helicase activity by TRF2. Stimulatory effect is calculated relative to the helicase activity without TRF2 (Lane 2 for DL, lane 8 for DL-Tg2). (B) Gel showing the effect of Tg lesions on shelterin protein binding to D-Loops. 25, 50 and 100 nM TRF1 or TRF2 was incubated with 0.5 nM DL (lanes 1-7) or DL-Tg2 (lanes 8-14) as described in the methods. The percent of substrate DL (white bars) and DL-Tg2 (black bars) bound by each protein was quantified over three repeats. (C) Gel and quantitative analysis showing the effect of TRF2 on RECQL4 binding to D-Loops, where 80 nM RECQL4 binding alone (lanes 2 and 6) are compared with the binding of 80 nM RECQL4 in the presence of 40 nM TRF2 (lanes 3 and 7) on DL and DL-Tg2 as indicated. The binding capacity of 40 nM TRF2 alone on these substrates is also shown (lanes 4 and 8). Error bars show standard deviation over at least three independent assays. *P<0.05 by a t-test.
