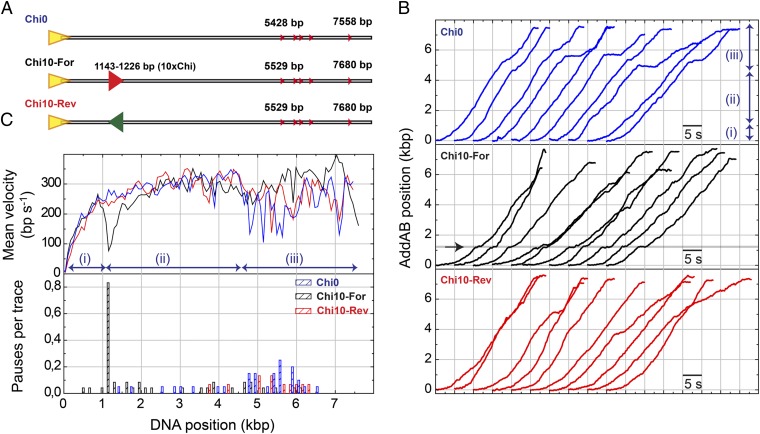
Fig. 2.
AddAB pauses at Chi sequences. (A) The standard DNA substrate Chi0 and DNA substrates containing 10 closely spaced Chi sequences (10×Chi) either correctly oriented for AddAB recognition (Chi10-For) or in opposite orientation (Chi10-Rev). The 10×Chi locus is located ∼1.1 kbp from the entry point. (B) Representative sets of wild-type AddAB translocation time traces for Chi0, Chi10-For, and Chi10-Rev. In the standard substrate (blue data) three regimes were identified: (i) arrival of ATP, (ii) constant AddAB velocity, and (iii) variable rates and pauses. A clear pause was found at the correctly oriented Chi locus (arrow in Chi10-For data). A control experiment using Chi10-Rev did not show a pause at Chi. (C) Mean AddAB velocity as a function of DNA position (Upper) and pauses per trace distribution for different DNA substrates (Lower) were calculated as described in SI Materials and Methods. About 80% of traces presented a clear pause of AddAB at the Chi locus.