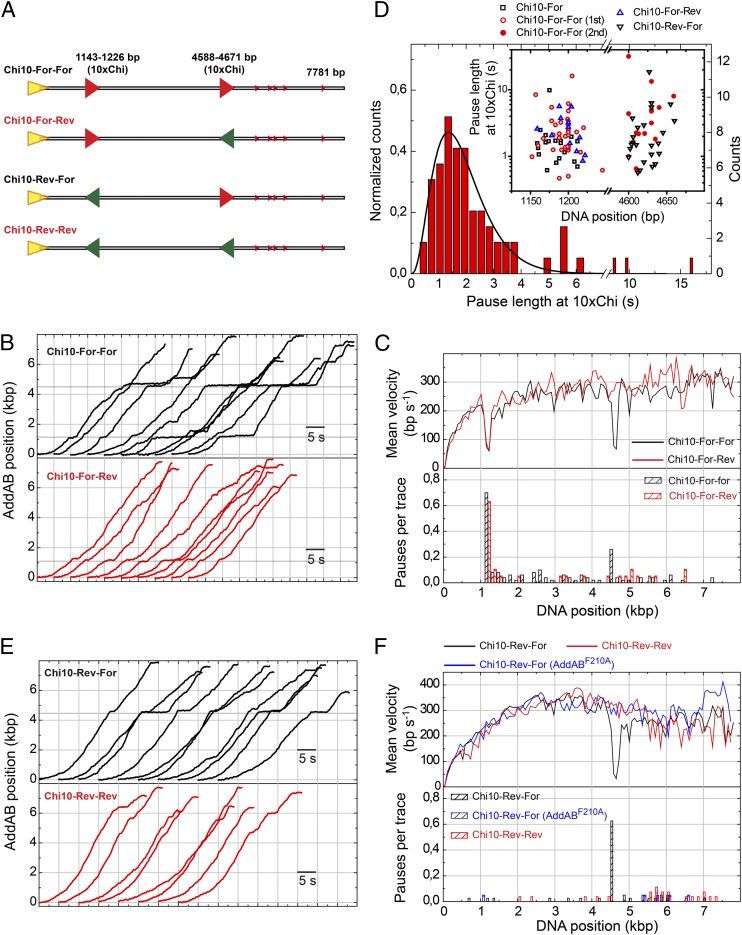
Fig. 3.
Conditional Chi recognition experiments and pause duration at Chi. (A) Cartoon representation of DNA substrates containing two Chi loci in all possible orientations (For-For, For-Rev, Rev-For, and Rev-Rev). (B) Representative sets of WTAddAB translocation time traces for the Chi10-For-For and Chi10-For-Rev substrates. Gray regions represent 10×Chi loci appropriately oriented for AddAB recognition. (C) Mean velocity graph and pause distribution histogram for wild-type AddAB translocation on Chi10-For-For and Chi10-For-Rev substrates, showing Chi-dependent pausing. (D) Normalized pause length distribution for all pauses at the first 10×Chi locus. Data were fitted to a normalized gamma function (SI Materials and Methods), giving N = 3.9 ± 0.3 and k = 2.1 ± 0.3 s−1. This distribution suggests a process made of multiple stochastic steps. (Inset) Pause position and duration at the first and second 10×Chi loci. No significant differences were found between pauses at the first and second loci in terms of their duration. (E) Representative sets of WTAddAB translocation time traces for the Chi10-Rev-For and Chi10-Rev-Rev substrates. (F) Mean velocity graph and pause distribution histogram for wild-type AddAB and AddABF210A translocation on Chi10-Rev-For and Chi10-Rev-Rev substrates. Data show that AddAB paused precisely at the Chi loci and implies that the positional accuracy of our measurements is ∼75 bp.