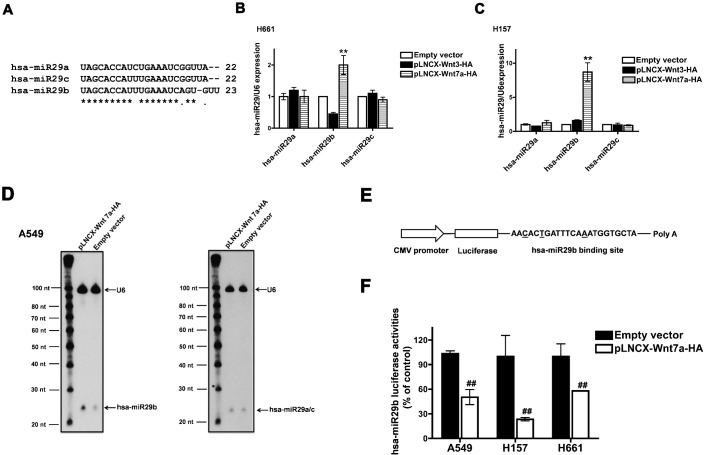
Fig. 1. Wnt7a/Fzd9 signaling regulates hsa-miR29b.
(A) Multiple alignments of hsa-miR29a, hsa-miR29b and hsa-miR29c. Real-time PCR analyses of the expression of hsa-miR29a, hsa-miR29b and hsa-miR29c in NSCLC cell lines. H661 (B) or H157 (C) cells were transfected either with empty vector, pLNCX-Wnt3-HA or pLNCX-Wnt7a-HA. After 24 h, total RNA was extracted and reverse transcribed. Real-time PCR analysis was carried out using the cDNAs and hsa-miR29a, hsa-miR29b or hsa-miR29c specific primers. RNU6B was used as the internal control for normalization. Data represent mean ± SEM of three separate experiments performed in duplicates. **P<0.01; versus empty vector control. (D) Northern blot analysis of Wnt7a induced expression of hsa-miR29a, hsa-miR29b, hsa-miR29c and U6 in A549 cells. A549 cells were transfected with either empty vector or pLNCX-Wnt7a-HA. Total RNA was extracted and low molecular weight RNA was enriched, and Northern analysis was performed as described in Materials and Methods. (E) Schematic representation of hsa-miR29b luciferase reporter plasmid. The binding site of hsa-miR29b differs from that of hsa-miR29a or hsa-miR29c at the underscored bases. (F) hsa-miR29b luciferase reporter assay in NSCLC cell lines. A549, H157 and H661 cells were transfected either with empty vector or pLNCX-Wnt7a-HA along with hsa-miR29b luciferase reporter plasmid. After 48 h, the cells were lysed and luciferase activities were measured as described in Materials and Methods. H157 cells were also transfected with Fzd9, as they do not express Fzd9. Luciferase values were normalized to CMV-β-galactosidase values were represented in the graph. Wnt7a-induced hsa-miR29b expression is represented as the percentage of empty vector control. Data represent mean ± SEM of three separate experiments. ##P<0.01; versus empty vector control.