Figure 1.
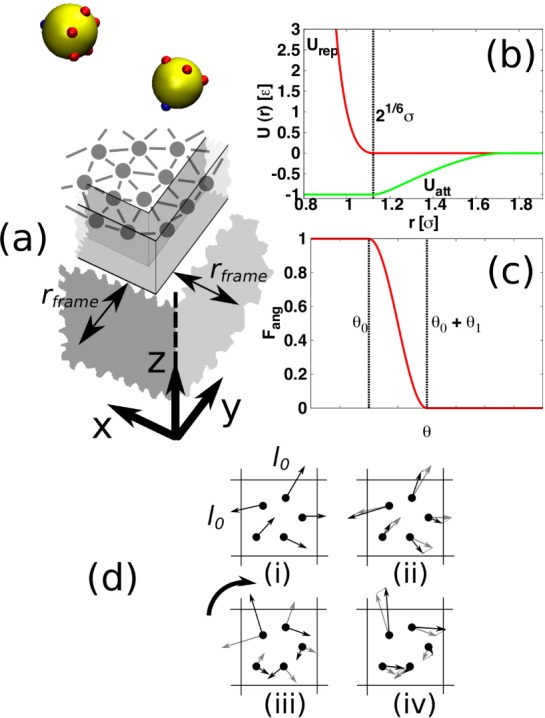
(a) Simulation setup. Subunits, which are all identical, are rendered in yellow, with positions, but not extents, of patches for interactions with other subunits in red. Positions of patches for interactions with the membrane particles are in blue. The membrane is modeled as a triangulated surface of bonded particles. The particles forming the surface edge are confined to a frame region, which is located at a distance rframe from the periodic boundaries. In simulations with hydrodynamics, a stochastic rotation dynamics (SRD) solvent composed of point particles is included. Interactions between SRD particles are effected by first dividing the entire system into a grid of cells of side l0. (b) The radial part of the inter-subunit or subunit-membrane potential, U(r), with a well-depth ε is split into attractive (green) and repulsive parts (red). (c) The attractive part is multiplied by factors of the form Fang(θ), where θ is an angle that depends on the relative orientation of the interacting particles. (d) Sketch of momentum transfer between SRD particles in a cell: (i) Only particles within one cell interact. (ii) Velocities are subtracted from all particles such that the center of mass velocity is 0. (iii) All velocities are rotated, as signified by the heavy arrow, around a random axis, by a given angle. (iv) The subtracted velocities are added back on so that total momentum is conserved.
