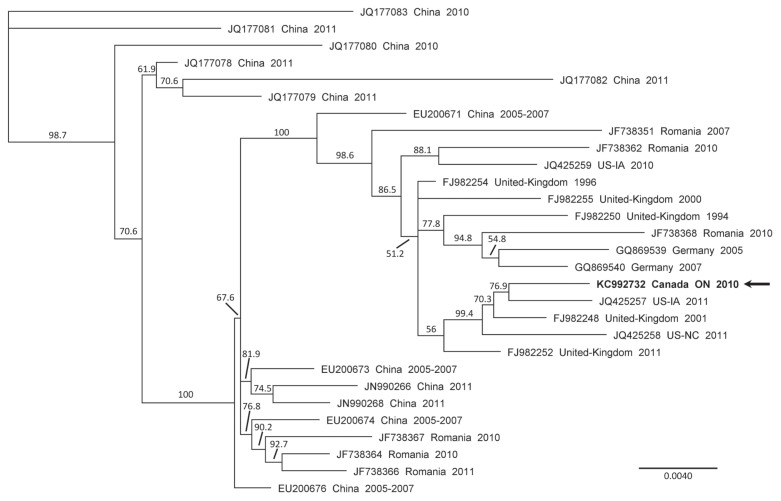
Figure 1.
Phylogenetic tree of the genomic nucleotide sequences of porcine partetravirus strains. The phylogenetic tree was generated by the neighbor-joining method using the Geneious V5.6.6 software with a bootstrap resampling method (1000 replications) using the longest common nucleotide sequences between all compared partetravirus genomic sequences. Bootstrap confidence levels are indicated at the nodes of the phylogenetic tree. GenBank accession numbers for the sequences of all viruses are provided as well as their countries of origin and their date of collection (year). The horizontal scale indicates the distances between strains; a distance of 0.0040 means that the strains possess 99.6% nucleotide identity. The arrow indicates the porcine partetravirus detected in the Ontario swine serum.