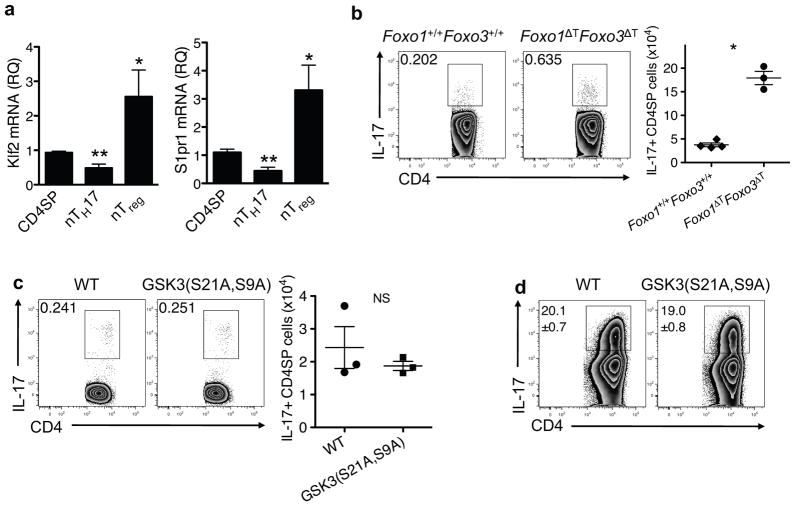
Figure 4.
Foxo proteins regulate nTH17 cell development. (a) The relative quantity (RQ) of Klf2 and S1pr1 mRNA transcripts in the indicated purified thymocyte populations, relative to β-actin, were determined by real-time PCR. CD4SP: CD4+CD44loCCR6−; nTH17: CD4+CD44hiCCR6+; nTreg: CD4+Foxp3+ from Foxp3-GFP reporter mice. Data are from 3 independently sorted thymic populations. All samples were run in triplicate; bars and error bars represent mean ± SEM. *P ≤ 0.05, **P ≤ 0.01 (one-way ANOVA followed by Dunnett’s post-test with CD4SP as control group). IL-17 producing thymocytes from Foxo1+/+Foxo3+/+ and Foxo1ΔTFoxo3ΔT mice (b) or WT and GSK3(S21A,S9A) mice (c) following ex vivo stimulation. Representative flow plots are gated on CD4SP TCRβ+TCRγδ− cells and the graph shows pooled data from two independent experiments (n = 3; mean ± SEM; NS, not significant; *P<0.0001, P value from two-tailed Student’s t-test). (d) Production of IL-17 from WT and GSK3(S21A,S9A) CD4+ T cells cultured with anti-CD3 plus anti-CD28 in iTH17-polarizing condition for 3 days followed by restimulation with PMA/ionomycin. Numbers represent mean ± SEM of IL-17+ cells from triplicate samples. Data are representative of three independent experiments.