Abstract
It is well established that catecholamine-stimulated thermogenesis in brown fat requires β-adrenergic elevations in cyclic AMP (cAMP) to increase expression of the uncoupling protein 1 (UCP1) gene. However, little is known about the downstream components of the signaling cascade or the relevant transcription factor targets thereof. Here we demonstrate that cAMP- and protein kinase A-dependent activation of p38 mitogen-activated protein kinase (MAPK) in brown adipocytes is an indispensable step in the transcription of the UCP1 gene in mice. By phosphorylating activating transcription factor 2 (ATF-2) and peroxisome proliferator-activated receptor gamma (PPARγ) coativator 1α (PGC-1α), members of two distinct nuclear factor families, p38 MAPK controls the expression of the UCP1 gene through their respective interactions with a cAMP response element and a PPAR response element that both reside within a critical enhancer motif of the UCP1 gene. Activation of ATF-2 by p38 MAPK additionally serves as the cAMP sensor that increases expression of the PGC-1α gene itself in brown adipose tissue. In conclusion, our findings illustrate that by orchestrating the activity of multiple transcription factors, p38 MAPK is a central mediator of the cAMP signaling mechanism of brown fat that promotes thermogenesis.
Brown adipose tissue (BAT), a thermogenic organ, is present in most mammals and is responsible for cold-induced nonshivering thermogenesis (reviewed in references 23, 27, 34, and 37). The distinctly russet tint of brown adipocytes derives from their densely packed mitochondria, within which resides a unique molecule known as uncoupling protein 1 (UCP1). In response to environmental challenges such as cold temperature or excess caloric intake, brown fat thermogenesis is triggered by a robust activation of the sympathetic nervous system (SNS), leading to a coordinated set of responses including brown adipocyte proliferation, mitochondrial biogenesis, and increased local blood flow to satisfy the augmented demands for respiration. At the cellular level, this program is set in motion by the β-adrenergic receptors (βARs) and the cyclic AMP (cAMP) signaling cascade (19). One of the key target genes activated by this SNS response is the UCP1 gene (reviewed in references 27 and 29). Since it is able to dissipate the mitochondrial proton gradient, which is normally converted into the global metabolic currency ATP, the resulting partial uncoupling renders metabolic reactions less efficient, the net result of which is an increased metabolic rate and the release of chemical energy as heat (reviewed in reference 10).
The UCP1 gene was first cloned in 1985 (1, 14), and studies of genetically manipulated mouse models have led to the general conclusion that it is the only protein of its kind capable of eliciting this classic nonshivering thermogenic response (27). At the molecular level, the regulatory promoter region of the UCP1 gene has now been studied in several species, defining a conserved region as a strong enhancer responsible for tissue-specific and cAMP-dependent expression (6, 18, 36). This enhancer contains a canonical peroxisome proliferator-activated receptor (PPAR)-responsive element (PPRE) and two putative cAMP-responsive elements (CREs), with various candidate transcription factors being proposed to regulate this enhancer region. The PPRE in the enhancer is conserved among species and has been shown to be essential, at least in cell culture models, for UCP1 enhancer activity (4, 36). The CRE sequences are not as well conserved. From mutagenesis studies in these in vitro models, we have learned that one of the two CREs in this enhancer region (CRE2) is likely important for norepinephrine-dependent stimulation of UCP1 gene transcription, while the other (CRE3) is dispensable (18). However, there has never been a clearly demonstrated role for CRE-binding protein (CREB) to regulate the UCP1 enhancer, although CREB can bind to the CRE in the proximal region of the rat UCP1 promoter (positions −139 to −122). This latter element alone is unable to activate the UCP1 gene but serves to augment the critical role of the UCP1 enhancer (45). Therefore, to date the mechanism(s) whereby cAMP signaling and nuclear factors intersect to coordinate the regulation of UCP1 expression is still unclear.
Signal transduction cascades emanating from G protein coupled receptors such as the βARs can recruit components of the growth factor kinase systems, creating a web of pathways coordinating transcriptional responses to hormonal and growth cues (reviewed in references 20 and 24). Similarly, advances in understanding the mechanisms of transcriptional regulation have included the discovery that complexes of nuclear coactivators and corepressors may themselves be targets of regulatory modifications such as phosphorylation that originate from transmembrane signaling (43). A molecule that increasingly appears to serve as a hub for transcriptional control of metabolic regulation is PPARγ coactivator 1 (now named PGC-1α). Originally isolated as a PPARγ-interacting protein from a brown fat cDNA library (33), PGC-1α has been shown to coordinate gene expression that stimulates mitochondrial biogenesis and oxidative metabolism in several cell types with relevance to metabolic fuel utilization (reviewed in reference 32). Recently, it has been shown that overexpression of the PGC-1α gene can promote the occurrence of brown fat features in human white adipocytes (39).
In this study, we demonstrate that p38 mitogen-activated protein kinase (MAPK) is a central regulator of the β-adrenergic and cAMP-dependent thermogenic response in brown fat. We identified two crucial targets of p38 MAPK. One is the nuclear factor activating transcription factor 2 (ATF-2), which is phosphorylated by p38 MAPK and coordinates the transcriptional induction of the UCP1 and PGC-1α genes through their CREs. The second target of p38 MAPK is the PGC-1α protein itself, which then drives transcription of the UCP1 gene through its association with PPARγ bound to the UCP1 promoter. Thus, this requirement for p38 MAPK to coordinate the recruitment of distinct transcriptional factors to the UCP1 promoter serves to precisely regulate this classic cAMP-dependent response in brown fat.
MATERIALS AND METHODS
Chemicals and plasmids.
SB202190 and SB203580 (hereafter referred to together as SB) and H89 were from Calbiochem. Forskolin (FSK) and isoproterenol (ISO) were from Sigma (St. Louis, Mo.). CL316243 (CL) was a gift from Elliot Danforth, Jr. of Wyeth-Ayerst. Antibodies against PPARγ were purchased from Santa Cruz Biotechnology, Inc. Antibodies against CREB and ATF-2 were from Cell Signaling Technology. The ATF-2 fragment was from New England Biolabs. The plasmid EN-Tk-CAT containing the UCP1 enhancer (positions −2530 to −2310) was constructed as previously described (4). The CRE2 mutant of the UCP1 enhancer was built by mutating the wild-type AGTCGTCA into AGTTGTCA (italics indicate the point of mutation). The PPRE mutant of the UCP1 enhancer was constructed by mutating the PPRE from −2490 TCACCC T TGACCA −2478 into TCACAA T TGACCA. The PGC-1α-CAT plasmid was constructed by cloning part of the PGC-1α promoter (positions −170 to +68) into a chloramphenicol acetyltransferase (CAT) expression vector. The construction of the expression vectors pECMV-PGC-1α (PGC-1α), pECMV-PGC-1αA3 (PGC-1αA3), pMSCV-PGC-1α, and pMSCV-PGC-1αA3 was described previously (31). The α18CRE-CAT, containing one copy (from positions −146 to −129) of the CRE from the human glycoprotein hormone α subunit gene, was described previously (7). CAT reporter vectors containing two copies of the PPRE from the acyl coenzyme A oxidase gene ([AOX]2-tk CAT) and expression vectors for human PPARγ and RXRα were gifts from Steven A. Kliewer. The expression vector of dominant negative p38α (pCDNA3-p38αAF) was a gift from J. Han (11). The expression vector for constitutively active MAPK kinase 6 (MKK6E) was a gift from Roger Davis. The construction of the β-actin-luciferase plasmid (β-actin-luc) was previously described (26).
Cell culture and transfection.
The HIB-1B brown preadipocytes (35) were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum. Cells were transfected with a total amount of plasmid DNA equal to 2.2 μg/well and 5 μl of Lipofectamine (Life Technologies) in six-well plates. As indicated, these DNA mixtures included EN-Tk-CAT (1.0 μg), [AOX]2-tk CAT, pCDNA3-p38αAF or PGC-1α-CAT, and β-actin-luc (0.2 μg). The PPARγ agonist rosiglitazone (1 μM) was added to the cells 12 h after transfection. Where indicated, HIB-1B cells were treated for 1 h with 5 μM SB202190 or 20 μM H89 prior to treatment with FSK (10 μM) for 6 h.
COS-7 cells were maintained in DMEM with 10% fetal bovine serum and transfected with a total amount of plasmid DNA equal to 2.2 μg/well and 5 μl of Lipofectamine in six-well plates. For testing the activities of EN-Tk-CAT, α18CRE-CAT, β2-CRE-CAT, or PGC-1α-CAT, COS-7 cells were transfected and treated with FSK (10 μM) for 6 h in the presence or absence of preincubation (1 h) of p38 MAPK inhibitor SB (5 μM). For the reconstitution of UCP1 enhancer activity, these DNA mixtures included EN-Tk-CAT (1.0 μg), pECMV-PGC-1α (0.5 μg), PPARγ (0.25 μg), RXRα (0.25 μg), and β-actin-luc (0.2 μg). Rosiglitazone (1 μM) was added to COS-7 cells at the initiation of the transfections.
BOSC cells were maintained in DMEM containing 10% cosmic calf serum (HyClone, Utah). The transfection of BOSC cells to produce recombinant retroviruses was performed with retrovirus expression vectors pMSCV-PGC-1α, pMSCV-PGC-1αA3, or pMSCV empty vector by calcium-phosphate coprecipitation and selected for neomycin resistance during two passages according to commercial recommendations (Clontech, Calif.). Retroviruses in the media were harvested and stored at −80°C until use.
Preparation of the primary brown adipocytes and retrovirus infections.
Primary brown adipocytes from C57BL/6J mice were prepared from interscapular BAT (IBAT) as described previously (4) and grown to 50% confluence in 100-mm-diameter dishes. Cells were infected with retroviruses expressing pMSCV-PGC-1α, pMSCV-PGC-1αA3, or pMSCV empty vector. Sixteen hours later, the infected cells were differentiated with insulin (50 nM), thyroid hormone (50 nM), and troglitazone (10 μM) for 10 days.
CAT and luciferase assays.
Cells were harvested to assay UCP1 enhancer activities 48 h after transfection. FSK (10 μM) was added for the last 6 h of the transfection to stimulate cAMP production, after which cell extracts were prepared in lysis buffer from a CAT enzyme-linked immunosorbent assay kit (Roche Molecular Biochemicals). CAT and luciferase assays were performed as previously described (25).
Western blotting.
Whole-cell lysates were prepared by rinsing cells with cold phosphate-buffered saline, followed by the addition of 2× Laemmli sample buffer. An aliquot (5 μg/well) was resolved with mini Tris-glycine gels (4 to 20%) (Invitrogen) and transferred to nitrocellulose membranes. The levels of phosphorylated and total p38 MAPKs were detected with a 1:1,000 dilution of each specific antiserum (9211S and 9212; New England Biolabs), followed by a 1:10,000 dilution of goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase (RPN5783; Amersham). The PGC-1α protein level was measured by Western blotting with antisera against PGC-1α (1:1,000) and goat anti-rabbit immunoglobulin G conjugated with alkaline phosphatase as the second antibody (RPN5783; Amersham). Fluorescent bands were visualized with a Storm PhosphorImager (Molecular Dynamics).
Protein kinase assay for p38 MAPK activity.
In vitro kinase assays were performed at 37°C for 30 min by using 10 μl of whole-cell lysates, 2 μg of substrate glutathione S-transferase-ATF-2 (1 to 109) (purchased from New England Biolabs), 250 μM ATP, and 10 μCi of [γ-32P]ATP in 40 μl of kinase reaction buffer. The ATF-2 fragment used in these experiments was a specific substrate for p38 MAPK (22). An equal amount of 2× Laemmli sample buffer was added to terminate the reactions. Proteins were resolved with 4 to 20% mini-gradient Tris-glycine gels (Invitrogen), and the level of protein phosphorylation was visualized by autoradiography. The level of p38 MAPK in the cell lysates was detected by Western blotting with antibodies against p38 MAPK.
RNA isolation and Northern blotting.
Total cellular RNA was prepared by the TRI reagent method according to the manufacturer's protocol (Molecular Research Center, Inc). For Northern blot analysis, RNA was denatured, fractionated through 1.2% agarose gels, and transferred to Biotrans nylon membranes (ICN) (8). Radiolabeled probes were prepared by random primer extension (Prime-It RmT; Stratagene) in the presence of [α-32P]dCTP to a specific activity of >2 × 109 cpm/μg of DNA. The UCP1 probe was a 300-bp BglI fragment (17), and the PGC-1α probe was the full length of the coding region of the PGC-1α gene. A rat cDNA probe for cyclophilin was used as an internal hybridization and quantification standard, and the blots were hybridized and washed, all as previously described (9, 38).
Animal experiments.
To measure the effect of cold exposure on p38 MAPK phosphorylation and expression of the UCP1 and PGC-1α genes, C57BL/6J mice (4 to 6 weeks old) were kept at thermoneutrality (28°C) for 3 days and then randomly divided into various groups. One group was kept at 28°C, and the rest of the animals were transferred to a 4°C environment for 0.5, 1, 3, 6, and 16 h, respectively, or for the other time indicated. Following euthanasia, the IBAT was rapidly collected and frozen in liquid nitrogen. A portion of the IBAT (about 1/3) was ground into powder and dissolved with 2× Laemmli sample buffer for Western blotting. The remaining 2/3 of the IBAT was used to recover RNA for Northern blotting as described above. Four independent animal experiments were performed.
To evaluate the effect of p38 MAPK inhibition on the expression of the UCP1 and PGC-1α genes, 18 male C57BL/6J mice (5 to 6 weeks old) were kept at a thermoneutral environment (28°C) for 3 days and then were divided into two groups for the following experiments. One group of mice was administered two intraperitoneal injections of SB203580 (12.5 mg/kg of body weight) dissolved in saline 16 h and 1 h before the cold exposure. The dose of SB was established in previous reports (2). Following treatment with SB or saline, the mice were placed at 4°C for 0 h, 1 h, or 4 h, with three mice per each time point. The control group of mice received two equivalent injections of saline. IBAT was harvested for kinase assays and Northern or Western blotting. Four independent experiments were performed.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed according to the commercialized manual (Upstate). Briefly, HIB-1B brown fat cells were differentiated for 5 days as previously described (4). Following differentiation, the cells were stimulated with vehicle solution or FSK (10 μM) for 6 h. For the cross-linking of transcriptional factors with genomic DNAs, cells were incubated with formaldehyde (1%) for 10 min. The cells were then washed, harvested with scrapers, and lysed with sodium dodecyl sulfate lysis buffer. The whole-cell lysates were sonicated to shear genomic DNAs to lengths of 200 to 1,000 bp. Lysates were centrifuged at 13,000 rpm (Eppendorf microcentrifuge) for 10 min, and the supernatants were collected. The supernatants were diluted 10-fold with ChIP dilution buffer in the presence of protease inhibitors. Salmon sperm DNA-50% protein A agarose slurry (80 μl) was added to the lysates, which were incubated for 30 min to reduce nonspecific background. Agarose beads were precipitated by brief centrifugation, and the supernatant was collected. The immunoprecipitation antibodies against PPREγ, ATF-2, or CREB were added to the 2-ml supernatant fraction and incubated overnight at 4°C with rotation. Sixty microliters of Salmon sperm DNA-Protein A agarose slurry was added, and the mixture was incubated for 1 h at 4°C with rotation to collect the antibody-histone-DNA complex. The presence of target DNA fragments was detected by PCR after the DNA was de-cross-linked with proteins. The primers used for the UCP1 enhancer were 5′-AGTGAAGCTTGCTGTCACTC-3′ and 5′-GTCTGAGGAAAGGGTTGACC-3′, while those for the CRE4-containing region were 5′-GAGTGACGCGCGGCTGGG-3′ and 5′-GGGCTAGGTAGTGCCAGT-3′. The primers for the PGC-1α promoter were 5′-ACTATAGGGCACGCGTGGTCGACG-3′ and 5′-ACACAGAGCACACACTCATGCAGG-3′.
RESULTS
Cold exposure stimulates p38 MAPK phosphorylation, PGC-1α expression, and UCP1 transcription in IBAT.
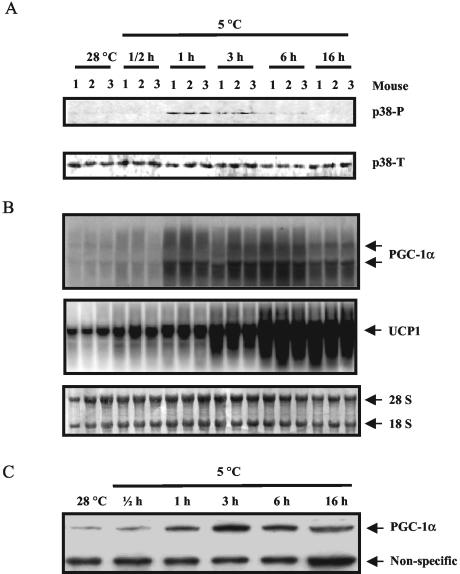
Cold exposure, through its activation of the SNS and the βARs in IBAT, is a strong stimulator of the expression of the UCP1 gene (reviewed in reference 27). We examined whether p38 MAPK could be activated in IBAT in vivo and its temporal relationship to the expression of the PGC-1α and UCP1 genes. As shown in Fig. 1A, in mice exposed to cold for up to 16 h, the level of phosphorylated p38 MAPK increased within a narrow time frame. Phospho-p38 was faintly visible at 30 min, peaked at 1 h (2.5-fold), and declined over the next few hours. Meanwhile, the increase of PGC-1α mRNA and protein became significant within 1 h (12-fold) (Fig. 1B and C), reached their maximal levels by 3 h (15-fold), and started to decline thereafter. UCP1 transcripts also increased in a time-dependent manner, reaching their maximum by 6 h (12-fold) and sustaining thereafter (Fig. 1B). Together, these data suggest a tight coordination between p38 MAPK activation and PGC-1α and UCP1 expression in IBAT during cold exposure.
FIG. 1.
Cold exposure promotes p38 MAPK phosphorylation, PGC-1α expression, and UCP1 transcription in IBAT. As described in Materials and Methods, IBAT was harvested from individual mice exposed to the cold to measure the following factors. (A) Phosphorylated p38 MAPK (p38-P) and total p38 MAPK (p38-T) in IBAT, measured by immunoblotting with specific antisera. (B) PGC-1α and UCP1 mRNA detected by Northern blotting. (C) PGC-1α protein assessed by immunoblotting with specific antisera; each lane represents the average of three mice per group. The results shown are from one of four independent experiments.
Activation of p38 MAPK is necessary for UCP1 expression in IBAT.
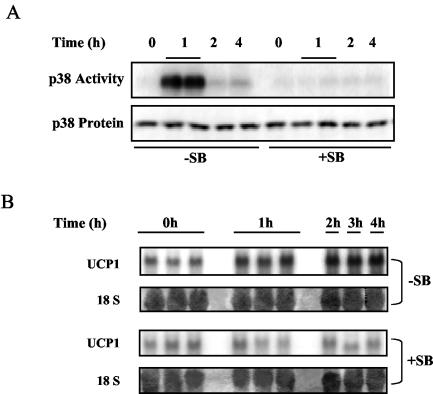
Since the results in Fig. 1 showed that p38 MAPK phosphorylation and UCP1 transcription may be related during cold exposure, an obvious question is whether p38 MAPK activation is required for the cold induction of the UCP1 gene. The SB series of p38α/β MAPK inhibitors has been widely used for experiments in vivo in both mice and rats (2, 16, 40, 41). Therefore, SB203580 was used to block the activation of p38 MAPK in mice in response to cold exposure, followed by an assessment of UCP1 expression levels in IBAT. As shown in Fig. 2A, p38 MAPK activity in IBAT, measured by the phosphorylation of a p38 MAPK-specific substrate, the ATF-2 fragment, was greatly increased by cold exposure, and this increase was totally abolished by pretreatment of the mice with SB. As a result, the cold induction of UCP1 gene expression in IBAT was completely prevented by the treatment with SB (Fig. 2B). Together, these data strongly support the notion that p38 MAPK activation is necessary for the cold induction of the UCP1 gene.
FIG. 2.
Cold activation of p38 MAPK is necessary for UCP1 expression in IBAT. As described in Materials and Methods, mice were treated with two doses of the p38 MAPK inhibitor SB203580 (+SB) or vehicle solution (−SB) at 16 and 1 h before the cold exposure. IBAT was harvested from each animal to measure the levels of p38 MAPK activity and UCP1 mRNA. (A) p38 MAPK activity measured by in vitro phosphorylation of the p38 MAPK specific substrate, the ATF-2 fragment, by IBAT lysates. Levels of p38 MAPK in the tissue lysates were measured by immunoblotting with antibodies against p38 MAPK. (B) UCP1 mRNA detected by Northern blotting. The level of 18S RNA was detected with methylene blue in the blot. All results are from one of four independent experiments.
cAMP- and p38 MAPK-dependent induction of the UCP1 gene requires both the PPRE and CRE2 of the UCP1 enhancer.
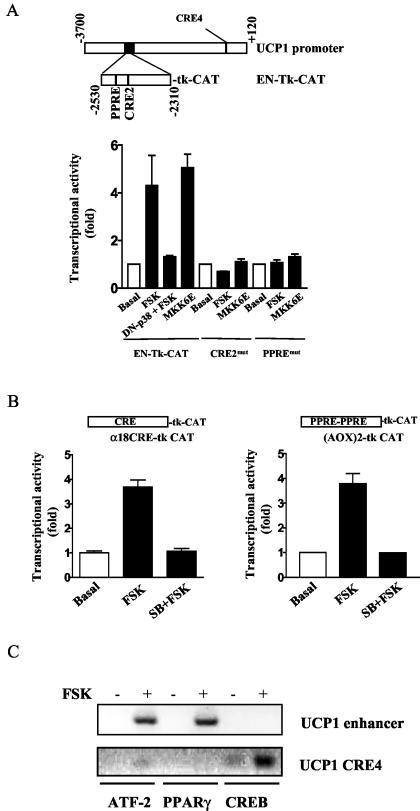
Scanning mutagenesis within an enhancer region of the UCP1 promoter that is conserved among species previously indicated two regions that were important for cAMP- or norepinephrine-dependent transcription (18). One of these elements was subsequently identified as a PPRE (36), and a second region was designated CRE2 as a putative CREB binding site (18). However, the identity of the transcription factors that bind these motifs and how they are controlled by cAMP have eluded detection. Since the UCP1 enhancer has been well defined to represent the majority of the adrenergic response and tissue-specific expression of the UCP1 gene (6, 18, 36), we compared the activities of either cAMP or p38 MAPK in transactivating a UCP1-enhancer-Tk-CAT reporter construct in HIB-1B brown adipocytes. Figure 3A shows that the adenylyl cyclase stimulator FSK increased UCP1 enhancer activity by 4.22-fold ± 1.20-fold (n = 3). This effect was completely blocked by the coexpression of a dominant negative inhibitory p38 MAPKα (p38αAF). On the other hand, the overexpression of a constitutively active form of MKK6 (MKK6E) stimulated UCP1 enhancer activity to a similar extent as did FSK (for MKK6E, 5.2-fold ± 0.35-fold; n = 3). Together, these results illustrate that the effects of either cAMP or p38 MAPK in increasing transcription of the UCP1 gene are both centered on components within the enhancer. Therefore, we next tested the effect of point mutations of the PPRE and CRE2 in the UCP1 enhancer on transcriptional activity elicited by FSK or MKK6E to determine whether these elements were independently regulated or required common signals and/or each other's functional integrity. Responses to both FSK and MKK6E were totally eliminated when either the PPRE or CRE2 was mutated.
FIG. 3.
(A) Both PPRE and CRE2 of the UCP1 enhancer are necessary for p38 MAPK-dependent activation of the UCP1 enhancer. As detailed in Materials and Methods, HIB-1B cells were transfected with wild-type UCP1 enhancer (EN-Tk-CAT) or the UCP1 enhancer containing a point mutation at either PPRE (PPREmut) or CRE2 (CRE2mut). As indicated, cells were either treated with FSK (10 μM) during the last 6 h of the transfection or cotransfected with an expression vector for dominant negative p38 MAPKα (p38αAF) or constitutively active MKK6 (MKK6E). The CAT activities were measured and normalized to β-actin luciferase. The results shown are means ± standard deviations of three independent experiments, each performed in duplicate. (B) p38 MAPK regulates the activities of the consensus PPRE and CRE in brown fat cells. The [AOX]2-tk CAT plasmid containing two copies of consensus PPRE or CRE (α18CRE) was introduced into HIB-1B cells via transient transfection. Cells were then treated with FSK for the last 6 h of the transfection in the absence or presence of incubation with SB. CAT activities were measured and normalized to β-actin luciferase. The results shown are means ± standard deviations of two independent experiments. (C) Both ATF-2 and PPARγ, but not CREB, bind to the UCP1 enhancer. HIB-1B cells were differentiated for 5 days as detailed in Materials and Methods. Cells were then stimulated with FSK for 6 h, followed by incubation with 1% formaldehyde to cross-link DNA and transcriptional factors and histones. DNA fragments sheared by sonication were precipitated with antibodies against ATF-2, CREB, or PPARγ, and the presence of the UCP1 enhancer and the proximal region of the UCP1 promoter containing the CRE4 in the precipitates was detected by PCR with specific primers. Results shown are representative of three independent experiments.
To determine whether p38 MAPK was capable of independently increasing the transcriptional activities of a PPRE and/or a CRE in brown adipocytes, we tested the ability of [AOX]2-tk CAT (21) or one copy of the CRE from the human glycoprotein hormone α subunit gene (α18CRE) (7) to respond to cAMP-elevating agents. As shown in Fig. 3B, FSK (10 μM) stimulated both [AOX]2-tk CAT (3.80-fold ± 0.41-fold) and α18CRE-CAT (2.61-fold ± 0.22-fold), and the p38 MAPK inhibitor SB (5 μM) completely blocked these responses. A series of constructs containing CREs from several other genes were equally sensitive to the inhibition of p38 MAPK when expressed in brown adipocytes, but in other cells types such as COS-7 and Hep-G2, there was no inhibition by SB (data not shown). This somewhat unexpected finding suggests that p38 MAPK may be a general mediator of cAMP signals to transcription factors interacting with PPREs and/or CREs in brown adipocytes.
To probe more directly for the transcription factors interacting with these promoter elements in the UCP1 enhancer, antibodies against PPARγ, phospho-ATF-2, or phospho-CREB were used to perform ChIP assays. As shown in Fig. 3C, antibodies against either PPARγ or ATF-2 could coprecipitate the UCP1 enhancer, but antibodies against CREB failed to do so. CREB antisera did, however, coprecipitate the proximal region of the UCP1 promoter containing CRE4. This region, although incapable of stimulating UCP1 transcription by itself, has been previously identified in a scanning mutagenesis experiment to be an important contributor to the activity of the UCP1 promoter (18). Together, these data indicate that PPARγ and ATF-2 are recruited to their respective elements (PPRE and CRE2) in the UCP1 enhancer by cAMP, while CREB interacts with CRE4 in the proximal region of the UCP1 promoter.
Phosphorylation of PGC-1α by p38 MAPK is essential for UCP1 induction.
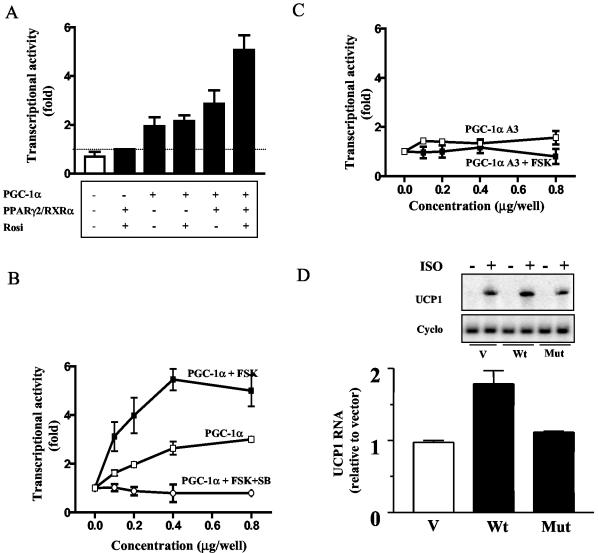
The nuclear coactivator PGC-1α was first isolated as a PPAR-interacting protein whose expression was increased in brown adipocytes in response to cAMP-elevating agents (33) and more recently was shown to be phosphorylated in skeletal muscle by cytokine activation of p38 MAPK (31). Therefore, we tested the hypothesis that PGC-1α was a critical target of β-adrenergic stimulated p38 MAPK activity in brown adipocytes to induce UCP1 expression via the PPRE. However, it was first necessary to reconstitute cAMP-inducible UCP1 enhancer activity in a cell background containing negligible levels of the prerequisite transcription factors for the expression of the UCP1 gene. Levels of endogenous PPARs and PGC-1α were very low in COS-7 cells (data not shown), and pilot experiments showed that when transfected alone, CAT activity from the UCP1 enhancer construct was undetectable under either basal or stimulated conditions. As shown in Fig. 4A, when expression vectors for PPARγ, RXRα, PGC-1α, and the PPARγ agonist rosiglitazone were sequentially introduced into the cells, maximal UCP1 enhancer activity was achieved by treatment with FSK. Parenthetically, PPARα was as effective as PPARγ in this reconstituted system (data not shown), a point that is relevant since there is a substantial amount of PPARα in brown adipocytes.
FIG. 4.
Phosphorylation of PGC-1α by p38 MAPK is necessary for cAMP-dependent UCP1 expression. (A) UCP1 enhancer activity reconstituted in COS-7 cells. COS-7 cells were cotransfected with EN-Tk-CAT and expression vectors of PGC-1α (0.5 μg/well), PPARγ (0.25 μg/well), and RXRα (0.25 μg/well). Rosiglitazone (1 μM) (Rosi) was added to the cells at the beginning of the transfection as noted. Cells were treated with FSK (10 μM) during the last 6 h of transfection. In panels B and C, COS-7 cells were similarly cotransfected with EN-Tk-CAT and the expression vectors of PPARγ, RXRα, and various amounts of wild-type PGC-1α. As indicated, some cells were treated with FSK with or without a preincubation with SB (B) or the PGC-1α, mutant PGC-1αA3 (C). The white bar in panel A represents the UCP1 enhancer activity in the absence of PPARγ/RXRα, rosiglitazone, PGC-1α, and FSK. In the experiments described above, CAT activities were measured in whole-cell lysates 48 h after the transfection and were normalized to luciferase. The results shown are means ± standard deviations of three independent experiments, each performed in duplicate. (D) PGC-1α stimulation of UCP1 gene transcription requires its phosphorylation by p38 MAPK in primary brown adipocytes. Cells were prepared as described in Materials and Methods, infected with empty retrovirus vector (V) or the vectors expressing wild-type pMSCV-PGC-1α (Wt) or the PGC-1α mutant pMSCV-PGC-1αA3 (Mut), and stimulated with βAR agonist ISO (10 μM) for 6 h. The transcripts of UCP1 and cyclophilin (Cyclo) were detected by Northern blotting. The results shown are means ± standard deviations of three independent experiments.
Using these conditions, we directly examined whether PGC-1α is a target of p38 MAPK and whether its phosphorylation is necessary for UCP1 enhancer activity. As shown in Fig. 4B, PGC-1α alone produced a modest concentration-dependent increase in UCP1 transcription. However, FSK produced a robust increase in UCP1 enhancer activity of 5.46-fold ± 0.46-fold over the basal level. Importantly, under these same conditions, a PGC-1α construct (PGC-1αA3) in which the p38 MAPK phosphorylation sites were mutated (T262A, S265A, and T298A) (31) was completely unable to promote transcription of the UCP1 enhancer under either basal or stimulated conditions (Fig. 4C). In addition, treatment with SB also completely prevented the induction of UCP1 enhancer activity by PGC-1α and FSK. While this result strongly supports the conclusion that the phosphorylation of PGC-1α is central to the cAMP-dependent control of UCP1, it was necessary to evaluate this conjecture in a more physiologic context. Primary brown adipocytes in culture were infected with retroviruses expressing wild-type PGC-1α, the mutant PGC-1αA3, or the empty vector, and the ability of the β-adrenergic agonist ISO to induce the expression of the endogenous UCP1 gene was measured. The results in Fig. 4D show that provision of wild-type PGC-1α augmented the β-adrenergic induction of the endogenous UCP1 gene by 100%, but the mutation of PGC-1α completely abolished this effect. Together, these data support a critical role for p38 MAPK phosphorylation of PGC-1α in the adrenergic stimulation of UCP1 gene transcription in brown adipocytes.
Activation of p38 MAPK is essential for cAMP-dependent transcription of the PGC-1α gene in IBAT.
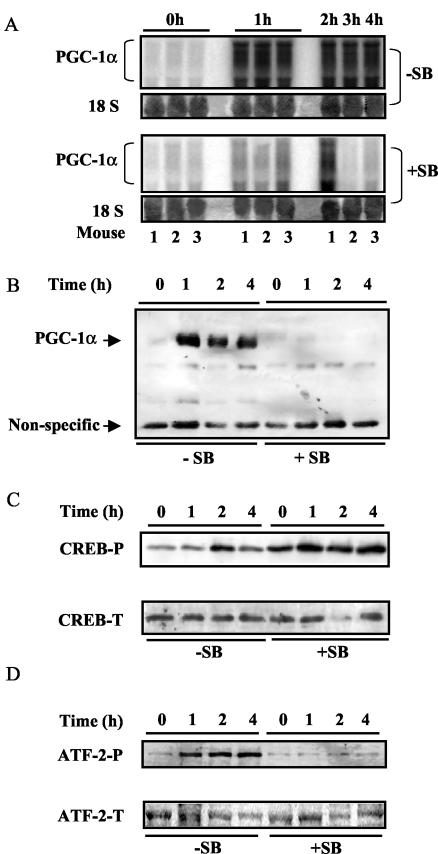
PGC-1α transcripts are increased in brown adipocytes by cold exposure or β-adrenergic stimulation (33). A perfect consensus CRE is present in the proximal promoter of the PGC-1α gene (−130 TGACGTCA −123), and the transcription factor CREB has been reported to be necessary for cAMP-dependent PGC-1α expression, at least in hepatocytes (13). Therefore, it was surprising to find that although PGC-1α mRNA and protein levels in IBAT were greatly elevated by cold exposure for 4 h (Fig. 5A and B), this response was completely abolished by pretreatment with the p38 MAPK inhibitor SB (Fig. 5A and B). This could have been a consequence of an SB-dependent loss in expression of CREB or of its phosphorylation. However, as shown in Fig. 5C, the overall levels of CREB were not different for the control and SB-treated groups. Moreover, upon cold exposure, CREB phosphorylation was increased in either the absence or presence of SB and was perhaps even slightly higher in the latter. On the other hand, the phosphorylation of ATF-2, another CRE-binding protein (28) and substrate of p38 MAPK, was totally blocked by the treatment in vivo with SB (Fig. 5D).
FIG. 5.
p38 MAPK is necessary for PGC-1α expression and differentially regulates the phosphorylation of CREB and ATF-2 in IBAT. As detailed in Materials and Methods, mice pretreated with the p38 MAPK inhibitor SB (+SB) or vehicle (−SB) were placed at 4°C for the time indicated. IBAT was harvested from individual mice to analyze the following factors by Northern or Western blotting. (A) PGC-1α mRNA and 18S RNA (same as in Fig. 2B). (B) PGC-1α protein. (C) Phosphorylated CREB (CREB-P) and total CREB (CREB-T). (D) Phosphorylated ATF-2 (ATF-2-P) and total ATF-2 (ATF-2-T). The results represent one of four independent experiments.
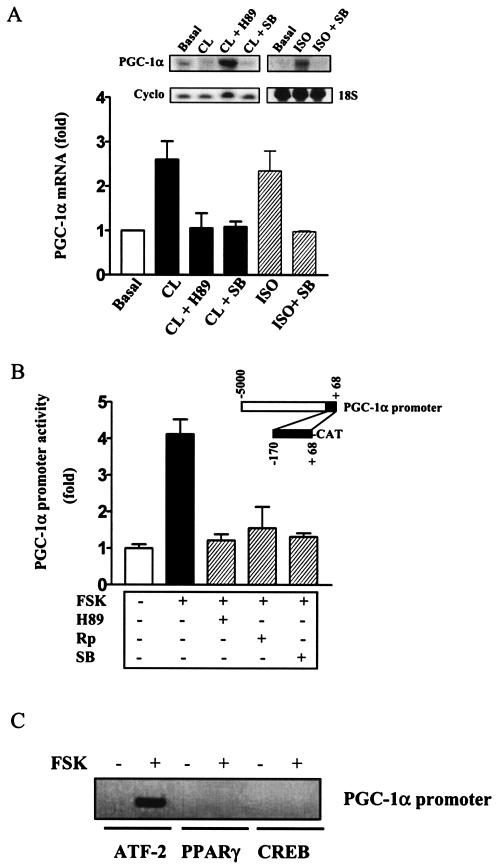
To determine whether p38 MAPK actually plays a direct role in the regulation of the PGC-1α gene in isolated brown adipocytes, the induction of PGC-1α mRNA by β-adrenergic agonists was examined in primary cultures of brown adipocytes in the presence of various kinase inhibitors. As shown in Fig. 6A, PGC-1α transcription was increased by either the β3AR agonist CL (2.62-fold ± 0.38-fold, n = 3) or the nonselective βAR agonist ISO (2.41-fold ± 0.42-fold, n = 3). As expected, this was completely prevented by the protein kinase A (PKA) inhibitor H89. In addition, β-agonist induction of PGC-1α mRNA was also blocked by p38 MAPK inhibitor SB, consistent with our results from IBAT in vivo (Fig. 5A and B). Furthermore, as shown in Fig. 6B, a PGC-1α-promoter (positions −170 to +68)-CAT reporter construct, which contains the sole functional CRE in the promoter (13), displayed the same response profile when expressed in HIB-1B brown adipocytes. The stimulation by FSK (4.10-fold ± 0.34-fold, n = 2) was completely blocked by the PKA inhibitors H89 and Rp-cAMP or by p38 MAPK inhibitor SB. Since both H89 and Rp-cAMP inhibited the response to FSK, this implicates a role for PKA but not other cAMP-dependent signaling elements such as Epac (4, 12) in the control of PGC-1α transcription. All together, these data clearly demonstrate that the cAMP-dependent increase in PGC-1α expression (33) depends not only on PKA (13) but also, at least in brown adipocytes, on p38 MAPK both in vitro and in vivo.
FIG. 6.
cAMP-dependent PGC-1α transcription in brown adipocytes is regulated by p38 MAPK. (A) Effect of the PKA inhibitor (H89) and p38 MAPK inhibitor (SB) on β-adrenergic stimulation of PGC-1α expression in primary brown adipocytes. Primary brown adipocytes were isolated and differentiated as described in Materials and Methods. Following incubation with CL (10 μM) or ISO (10 μM) for 6 h in the absence or presence of pretreatment (1 h) with H89 (20 μM) or SB (5 μM) as indicated, RNA was recovered from the cells to measure PGC-1α and cyclophilin (Cyclo) transcripts. The level of 18S RNA was visualized by staining with methylene blue in the blot as noted. (B) Effect of PKA and p38 MAPK inhibitors on the activity of the PGC-1α promoter. HIB-1B cells were transfected with PGC-1α-CAT vector as described in Materials and Methods. Cells were treated with FSK (10 μM) for the last 6 h of transfection with or without a preexposure to H89 (20 μM), Rp-cAMP (Rp) (1 mM), or SB (5 μM) for 1 h. The CAT activities were measured and normalized to luciferase. The results shown are means ± standard deviations of two independent experiments, each performed in triplicate. (C) ATF-2 binds to the promoter region of PGC-1α containing CRE. HIB-1B cells were differentiated for 5 days as detailed in Materials and Methods. Cells were then stimulated with FSK for 6 h, followed by incubation with 1% formaldehyde to cross-link DNA and transcriptional factors and histones. DNA fragments sheared by sonication were precipitated with antibodies against ATF-2, CREB, or PPARγ, and the presence of the proximal region of the PGC-1α promoter in the precipitates was detected by PCR with specific primers. Results shown are representative of three independent experiments.
To determine the transcription factors involved in the cAMP-dependent activation of the PGC-1α gene, ChIP assays were performed with antibodies against PPARγ, phospho-ATF-2, or phospho-CREB. As shown in Fig. 6C, this proximal region of the PGC-1α promoter was coprecipitated with ATF-2 but not by antisera to PPARγ or CREB. These results, together with the in vivo data from Fig. 5, indicate that ATF-2, but not CREB, controls PGC-1α gene transcription in brown adipocytes in response to cAMP.
DISCUSSION
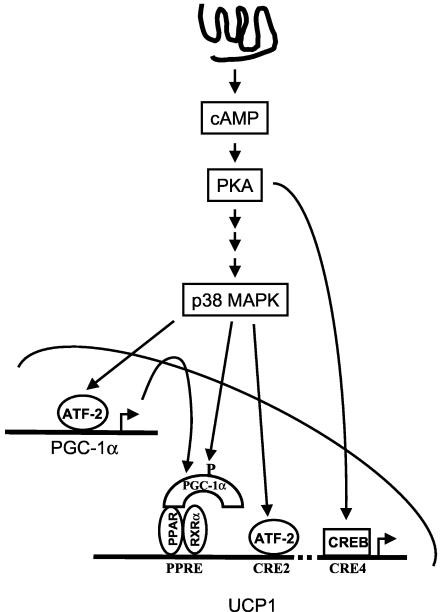
Among the coordinated cellular events triggered by catecholamines to elicit nonshivering thermogenesis in brown fat, the increased transcription of the UCP1 gene has a clear dependence on β-adrenergic activation of cAMP production (see references 27 and 29 for review), but the components of the signaling cascade and the transcription factors beyond this point have been ambiguous. Our findings from this study, together with previous observations (4), identify p38 MAPK as a central obligatory component of the signaling cascade mediating β-adrenergic regulation of UCP1 expression in BAT and cold acclimation. By a combination of in vivo and in vitro approaches, we showed that p38 MAPK regulates UCP1 gene transcription through a coordinated activation of nuclear factors on two separate elements (PPRE and CRE2) of the UCP1 enhancer region (Fig. 7). Specifically, cAMP-regulated transactivation of the UCP1 promoter requires p38 MAPK phosphorylation of both ATF-2 and PGC-1α, while CREB also plays an important role in a PKA-dependent, but p38-independent, manner. Finally, we showed that in addition to its acute activation of PGC-1α by phosphorylation, p38 MAPK also conveys the cAMP signal for increasing the overall expression of PGC-1α to further enhance mitochondrial thermogenic capability (42).
FIG. 7.
Schematic diagram of the signaling pathway of UCP1 transcription in brown fat.
From several different approaches, we demonstrated that the ability of p38 MAPK to control UCP1 gene transcription in brown adipocytes utilizes PGC-1α as a direct target, and this is accomplished at two levels: phosphorylation of the PGC-1α protein as well as an increase in its overall level of expression. Puigserver et al. previously showed that one of the underlying mechanisms for increased skeletal muscle mitochondrial respiration by inflammatory cytokines was phosphorylation of PGC-1α by p38 MAPK (31). Here we demonstrated that in brown adipocytes, this same phosphorylation of PGC-1α by p38 MAPK is also necessary for driving UCP1 gene transcription, but in this case the stimulus is cAMP and PKA. While the ability of cytokines such as tumor necrosis factor alpha or the interleukins to activate the p38 MAPK signaling cascade is fairly well established (see reference 15 for review), the process whereby PKA ultimately triggers p38 MAPK represents a new pathway that is currently the focus of our efforts. Thus, although the initial stimulus for p38 MAPK activation is different for brown adipocytes and myocytes, the net effect on PGC-1α coactivator function is the same (see Fig. 4 and reference 31). On the contrary, the transcriptional control of the PGC-1α gene is different among tissues. Expression of the PGC-1α gene in adipocytes was originally shown to be under the control of β-adrenergic stimulation and cAMP (33), and the transcription factor(s) mediating this response was presumed to be CREB acting downstream of PKA. Instead, our studies show that in brown adipocytes, the induction of PGC-1α by cAMP requires p38 MAPK. Using in vitro and in vivo experiments, we demonstrated that the downstream effector of p38 MAPK in brown adipocytes is ATF-2 rather than CREB. Therefore, the manner by which PGC-1α is regulated in brown adipocytes contrasts with the mechanism in hepatocytes, where CREB phosphorylation is essential (44).
We also show that ATF-2 not only regulates the expression of the PGC-1α gene but also is directly involved in the control of UCP1 gene transcription. Using mutagenesis studies and ChIP assays, we showed that both the PPRE and CRE2 elements in the UCP1 enhancer region are required for cAMP- and p38 MAPK-dependent transcription, consistent with earlier studies that showed these two regions were required for the norepinephrine stimulation of the UCP1 promoter (18, 36). However, contrary to a general assumption that CREB is the vital mediator of β-adrenergic stimulation of UCP1 transcription, our results indicate that ATF-2, but not CREB, is required to regulate the critical enhancer region of the UCP1 promoter (5, 18, 36). In addition, we show that as an essential player, p38 MAPK regulates and coordinates the activities of the factors bound to both the PPRE and CRE2 by driving the expression and activity of PGC-1α and ATF-2 activation, respectively. One model based upon these results is that the activities of the PPRE and CRE2 are independently sensitive to p38 MAPK, in similarity to a safe-deposit box that requires two separate keys to open. Alternatively, since PGC-1α has been shown to increase transcriptional activity on a model promoter through the assembly of a complex that includes the histone acetyltransferase steroid receptor coactivator 1 and CREB/p300 (30), we do not exclude the possibility of a more intimate association between PGC-1α and ATF-2 that precisely regulates UCP1 expression, and these aspects are under investigation. Finally, using ChIP studies, we provide evidence that CREB indeed contributes to the cAMP responsiveness of the UCP1 gene by binding the CRE (presumably CRE4) at the proximal region of the promoter. This observation is consistent with a previous report that CREB can interact with the equivalent region of the rat UCP1 promoter (45).
Many key features of the brown fat thermogenic response have been shown to be dependent on β-adrenergic stimulation of cAMP production, including proliferation (3), UCP1 and type II deiodinase expression, mitochondrial biogenesis, and respiration (27). Whether all these events are regulated by p38 MAPK in the same way as UCP1 and PGC-1α expression in brown fat should now be fully explored.
Acknowledgments
We thank Steve Kliewer for the expression vectors for PPARγ, RXRα, and [AOX]2-tk CAT. We thank J. Han for pCDNA3-p38αAF and Roger Davis for the expression vector of the constitutively active MKK6E.
This work was supported by NIH awards R01-DK53092 (S.C.), R01-DK54024 (S.C.), R01-DK54477 (B.M.S.), and Fonds de la Recherche en Santé du Québec (J.R.).
REFERENCES
- 1.Aquila, H., T. A. Link, and M. Klingenberg. 1985. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 4:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, A. M., J. N. Bradbeer, B. Votta, J. C. Lee, J. L. Adams, and D. E. Griswold. 1996. Pharmacological profile of SB 203580, a selective inhibitor of cytokine suppressive binding protein/p38 kinase, in animal models of arthritis, bone resorption, endotoxin shock and immune function. J. Pharmacol. Exp. Ther. 279:1453-1461. [PubMed] [Google Scholar]
- 3.Bukowiecki, L. J., A. Geloen, and A. J. Collet. 1986. Proliferation and differentiation of brown adipocytes from interstitial cells during cold acclimation. Am. J. Physiol. 250:C880-C887. [DOI] [PubMed] [Google Scholar]
- 4.Cao, W., A. V. Medvedev, K. W. Daniel, and S. Collins. 2001. β-Adrenergic activation of p38 MAP kinase in adipocytes. cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J. Biol. Chem. 276:27077-27082. [DOI] [PubMed] [Google Scholar]
- 5.Cassard-Doulcier, A. M., C. Gelly, F. Bouillaud, and D. Ricquier. 1998. A 211-bp enhancer of the rat uncoupling protein-1 (UCP-1) gene controls specific and regulated expression in brown adipose tissue. Biochem. J. 333:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassard-Doulcier, A.-M., C. Gelly, N. Fox, J. Schrementi, S. Raimbault, S. Klaus, C. Forest, F. Bouillard, and D. Ricquier. 1993. Tissue-specific and β-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5′-flanking region. Mol. Endocrinol. 7:497-506. [DOI] [PubMed] [Google Scholar]
- 7.Collins, S., J. Altschmied, O. Herbsman, M. G. Caron, P. L. Mellow, and R. J. Lefkowitz. 1990. A cAMP response element in the β2-adrenergic receptor gene confers transcriptional autoregulation by cAMP. J. Biol. Chem. 265:19330-19335. [PubMed] [Google Scholar]
- 8.Collins, S., M. G. Caron, and R. J. Lefkowitz. 1988. β2-Adrenergic receptors in hamster smooth muscle cells are transcriptionally regulated by glucocorticoids. J. Biol. Chem. 263:9067-9070. [PubMed] [Google Scholar]
- 9.Collins, S., K. W. Daniel, E. M. Rohlfs, V. Ramkumar, I. L. Taylor, and T. W. Gettys. 1994. Impaired expression and functional activity of the β3- and β1-adrenergic receptors in adipose tissue of congenitally obese (C57BL/6J ob/ob) mice. Mol. Endocrinol. 8:518-527. [DOI] [PubMed] [Google Scholar]
- 10.Dalgaard, L. T., and O. Pedersen. 2001. Uncoupling proteins: functional characteristics and role in the pathogenesis of obesity and type II diabetes. Diabetologia 44:946-965. [DOI] [PubMed] [Google Scholar]
- 11.Derijard, B., J. Raingeaud, T. Barrett, I. H. Wu, J. Han, R. J. Ulevitch, and R. J. Davis. 1995. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267:682-685. [DOI] [PubMed] [Google Scholar]
- 12.de Rooij, J., F. J. Zwartkruis, M. H. Verheijen, R. H. Cool, S. M. Nijman, A. Wittinghofer, and J. L. Bos. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396:474-477. [DOI] [PubMed] [Google Scholar]
- 13.Herzig, S., F. Long, U. S. Jhala, S. Hedrick, R. Quinn, A. Bauer, D. Rudolph, G. Schutz, C. Yoon, P. Puigserver, B. Spiegelman, and M. Montminy. 2001. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179-183. [DOI] [PubMed] [Google Scholar]
- 14.Jacobsson, A., U. Stadler, M. A. Glotzer, and L. P. Kozak. 1985. Mitochondrial uncoupling protein from mouse brown fat. J. Biol. Chem. 260:16250-16254. [PubMed] [Google Scholar]
- 15.Johnson, G. L., and R. Lapadat. 2002. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298:1911-1912. [DOI] [PubMed] [Google Scholar]
- 16.Joyeux, M., A. Boumendjel, R. Carroll, C. Ribuot, D. Godin-Ribuot, and D. M. Yellon. 2000. SB 203580, a mitogen-activated protein kinase inhibitor, abolishes resistance to myocardial infarction induced by heat stress. Cardiovasc. Drugs Ther. 14:337-343. [DOI] [PubMed] [Google Scholar]
- 17.Kozak, L., J. Britton, U. Kozak, and J. Wells. 1988. The mitochondrial uncoupling protein gene. J. Biol. Chem. 263:12274-12277. [PubMed] [Google Scholar]
- 18.Kozak, U. C., J. Kopecky, J. Teisinger, S. Enerback, B. Boyer, and L. P. Kozak. 1994. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol. Cell. Biol. 14:59-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafontan, M., and M. Berlan. 1993. Fat cell adrenergic receptors and the control of white and brown fat cell function. J. Lipid Res. 34:1057-1091. [PubMed] [Google Scholar]
- 20.Lefkowitz, R. J. 1998. G protein-coupled receptors. III. New roles for receptor kinases and β-arrestins in receptor signaling and desensitization. J. Biol. Chem. 273:18677-18680. [DOI] [PubMed] [Google Scholar]
- 21.Lehmann, J. M., L. B. Moore, T. A. Smith-Oliver, W. O. Wilkison, T. M. Willson, and S. A. Kliewer. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for the nuclear receptor PPARγ. J. Biol. Chem. 270:12953-12956. [DOI] [PubMed] [Google Scholar]
- 22.Li, Z., Y. Jiang, R. J. Ulevitch, and J. Han. 1996. The primary structure of p38 gamma: a new member of p38 group of MAP kinases. Biochem. Biophys. Res. Commun. 228:334-340. [DOI] [PubMed] [Google Scholar]
- 23.Lowell, B. B., and B. M. Spiegelman. 2000. Towards a molecular understanding of adaptive thermogenesis. Nature 404:652-660. [DOI] [PubMed] [Google Scholar]
- 24.Luttrell, L. M. 2002. Big g, little g. G proteins and actin cytoskeletal reorganization. Mol. Cell 9:1152-1154. [DOI] [PubMed] [Google Scholar]
- 25.Medvedev, A. V., S. K. Snedden, S. Raimbault, D. Ricquier, and S. Collins. 2001. Transcriptional regulation of the mouse uncoupling protein-2 gene: double E-box motif is required for PPARγ-dependent activation. J. Biol. Chem. 276:10817-10823. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto, N. G. 1987. Nucleotide sequence of the human beta-actin promoter 5′ flanking region. Nucleic Acids Res. 15:9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nedergaard, J., V. Golozoubova, A. Matthias, A. Asadi, A. Jacobsson, and B. Cannon. 2001. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochim. Biophys. Acta 1504:82-106. [DOI] [PubMed] [Google Scholar]
- 28.Ono, K., and J. Han. 2000. The p38 signal transduction pathway: activation and function. Cell. Signal. 12:1-13. [DOI] [PubMed] [Google Scholar]
- 29.Pecqueur, C., E. Couplan, F. Bouillaud, and D. Ricquier. 2001. Genetic and physiological analysis of the role of uncoupling proteins in human energy homeostasis. J. Mol. Med. 79:48-56. [DOI] [PubMed] [Google Scholar]
- 30.Puigserver, P., G. Adelmant, Z. Wu, M. Fan, J. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286:1368-1371. [DOI] [PubMed] [Google Scholar]
- 31.Puigserver, P., J. Rhee, J. Lin, Z. Wu, J. Yoon, C. Zhang, S. Krauss, V. Mootha, B. Lowell, and B. Spiegelman. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPAR coactivator-1. Mol. Cell 8:971. [DOI] [PubMed] [Google Scholar]
- 32.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24:78-90. [DOI] [PubMed] [Google Scholar]
- 33.Puigserver, P., Z. Wu, C. Park, R. Graves, M. Wright, and B. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829-839. [DOI] [PubMed] [Google Scholar]
- 34.Ricquier, D., and F. Bouillaud. 2000. The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J. 345:161-179. [PMC free article] [PubMed] [Google Scholar]
- 35.Ross, S. R., L. Choy, R. A. Graves, N. Fox, V. Solevjeva, S. Klaus, D. Ricquier, and B. M. Speigelman. 1992. Hibernoma formation in transgenic mice and isolation of a brown adipocyte cell line expressing the uncoupling protein gene. Proc. Natl. Acad. Sci. USA 89:7561-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears, I. B., M. A. MacGinnitie, L. G. Kovacs, and R. A. Graves. 1996. Differentiation-dependent expression of the brown adipocyte uncoupling protein gene: regulation by peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 16:3410-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silva, J., and R. Rabelo. 1997. Regulation of the uncoupling protein gene expression. Eur. J. Endocrinol. 136:251-264. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, P. S. 1980. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. USA 77:5201-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiraby, C., G. Tavernier, C. Lefort, D. Larrouy, F. Bouillaud, D. Ricquier, and D. Langin. 2003. Acquirement of brown fat cell features by human white adipocytes. J. Biol. Chem. 278:33370-33376. [DOI] [PubMed] [Google Scholar]
- 40.Underwood, D. C., R. R. Osborn, S. Bochnowicz, E. F. Webb, D. J. Rieman, J. C. Lee, A. M. Romanic, J. L. Adams, D. W. Hay, and D. E. Griswold. 2000. SB 239063, a p38 MAPK inhibitor, reduces neutrophilia, inflammatory cytokines, MMP-9, and fibrosis in lung. Am. J. Physiol. Lung Cell Mol. Physiol. 279:L895-L902. [DOI] [PubMed] [Google Scholar]
- 41.Underwood, D. C., R. R. Osborn, C. J. Kotzer, J. L. Adams, J. C. Lee, E. F. Webb, D. C. Carpenter, S. Bochnowicz, H. C. Thomas, D. W. Hay, and D. E. Griswold. 2000. SB 239063, a potent p38 MAP kinase inhibitor, reduces inflammatory cytokine production, airways eosinophil infiltration, and persistence. J. Pharmacol. Exp. Ther. 293:281-288. [PubMed] [Google Scholar]
- 42.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115-124. [DOI] [PubMed] [Google Scholar]
- 43.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 44.Yoon, J. C., P. Puigserver, G. Chen, J. Donovan, Z. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, C. B. Newgard, and B. M. Spiegelman. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131-138. [DOI] [PubMed] [Google Scholar]
- 45.Yubero, P., M. J. Barbera, R. Alvarez, O. Vinas, T. Mampel, R. Iglesias, F. Villarroya, and M. Giralt. 1998. Dominant negative regulation by c-Jun of transcription of the uncoupling protein-1 gene through a proximal cAMP-regulatory element: a mechanism for repressing basal and norepinephrine-induced expression of the gene before brown adipocyte differentiation. Mol. Endocrinol. 12:1023-1037. [DOI] [PubMed] [Google Scholar]