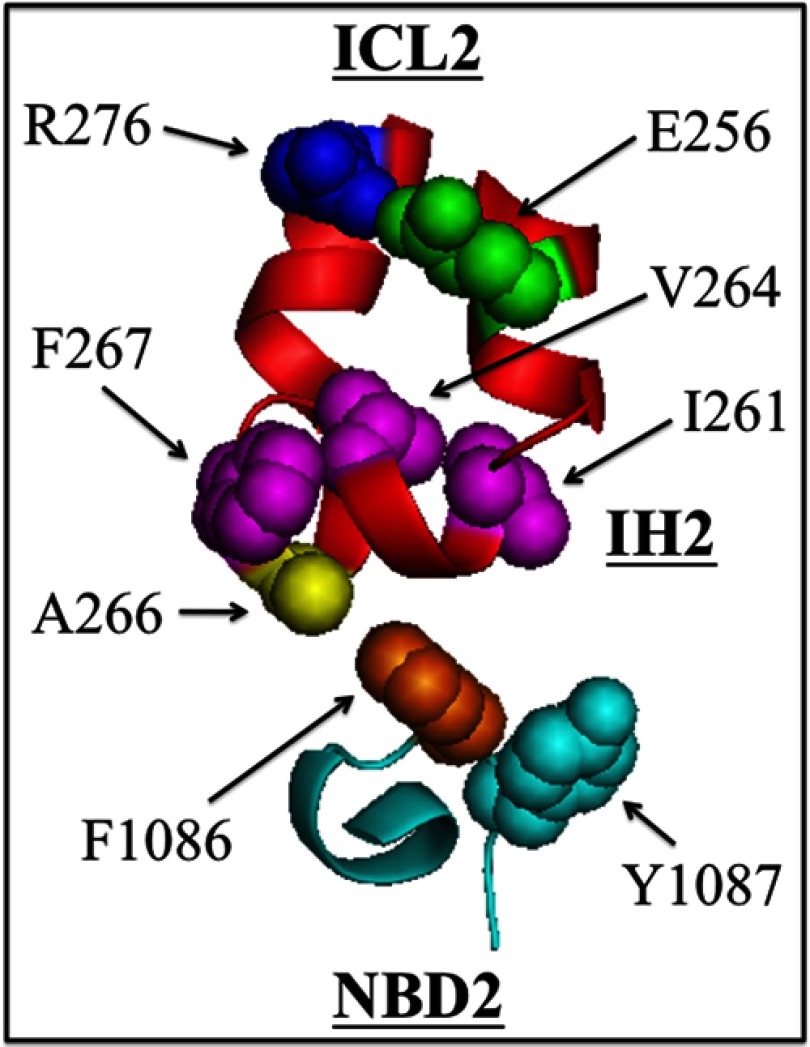
FIGURE 7.
Model showing hydrophobic amino acids at the IH2-NBD2 interface critical for P-gp maturation and/or activity. Residues Val-253–Asn-278 (ICL2) and Val-1080–Pro-1089 (NBD2) are shown. The locations of residues Ile-261, Val-264, Phe-267, and Tyr-1087 are shown as mutation to these residues inhibited maturation. Removal of the aromatic side chain of Phe-1086 (F1086A mutation) inhibited P-gp activity because it inhibited coupling between the ATP- and drug-binding domains. Replacement of Ala-266 with aromatic residues restored the activity of F1086A. Residues Glu-256 and Arg-276 were recently shown to form a salt bridge that was critical for P-gp maturation (22).