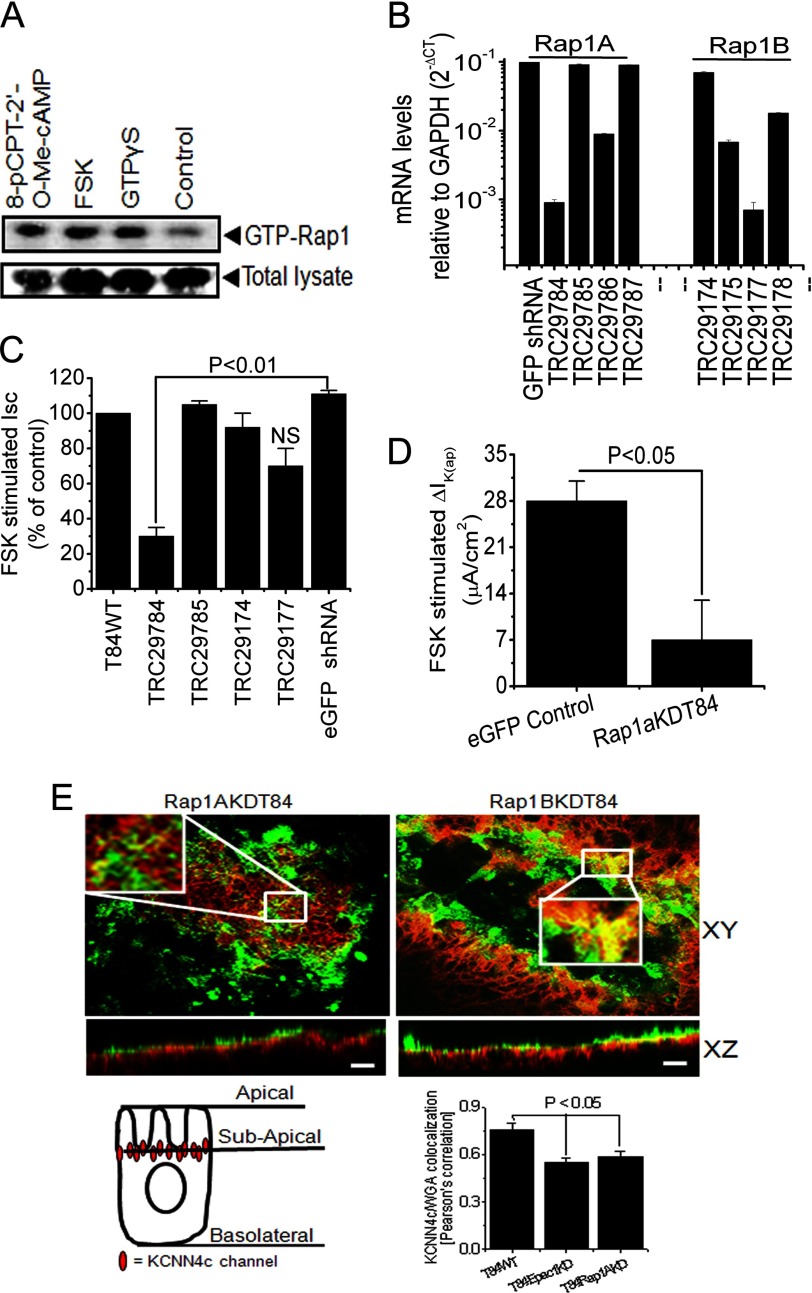
FIGURE 5.
Involvement of Rap1 in FSK-stimulated Isc and IK(ap) through Epac1 stimulation. A, endogenous Rap1 activity was measured with FSK or 8-pCPT-2′-O-Me-cAMP or without (control). Cells were incubated in serum-free medium. FSK and 8-pCPT-2′-O-Me-cAMP lead to an activation of endogenous Rap1 in T84WT cells. Western blots of total cell lysate probed for total Rap1 are shown as control. Cells loaded with GTPγS and incubated with RalGDS-RBD-agarose beads served as a positive control. A representative Western blot of three independent experiments is shown. B, quantitative PCR analysis of Rap1A and Rap1B expression in T84WT cells after lenti-shRNA transduction specific for Rap1A and Rap1B genes, which are shown as mRNA levels relative to levels of GAPDH in log scale. CT, cycle threshold. Values are mean ± S.E.; n = 3. Summary of the effects of Rap1A and Rap1B knockdown by gene-specific lenti-shRNA on FSK-stimulated Isc (C) and IK(ap) (D) of Rap1AKDT84 cells. FSK-stimulated Isc in T84WT cells was set to 100% (control). Enhanced GFP (eGFP) shRNA-transduced cell monolayers were used as a non-targeting control. Values are mean ± S.E.; n = 4. NS, statistically not significant. E, depletion of Rap1A affected the colocalization of KCNN4c with WGA on the apical membrane as reflected by a significant loss of yellow signal in the merged composite (xy and xz planes; top left versus top right). Insets show enlarged regions within the white box. Confluent Rap1AKDT84 and Rap1BKDT84 cell monolayers were stained with FITC-WGA on ice, then fixed, and stained for KCNN4c (red), and a stack of xz images was collected by confocal microscopy. A representative xz section is shown here. The scale bar represents 10 μm. Bottom right, quantification of KCNN4c-colocalized WGA in T84WT versus Epac1KDT84 or Rap1AKDT84 cells presented as mean ± S.E. by calculating the Pearson's colocalization coefficient using Carl Zeiss LSM software (version 3.2). The cell illustration (bottom left) indicates the redistribution of KCNN4c beneath the subapical membrane due to depletion of Epac1 or Rap1A. Error bars represent S.E.