Background: BAG6 complex functions in tail-anchored targeting and ERAD, but its role in the nucleus is not clear.
Results: BAG6 phosphorylation and nuclear localization are critical for DDR; UBL4A and GET4 translocate to nucleus upon DNA damage.
Conclusion: All three components participate in DDR.
Significance: Our work identified a nuclear function for the BAG6 complex.
Keywords: Cell Cycle, Cell Death, Checkpoint Control, DNA Damage Response, Protein Translocation, ATM, BAG6, GET4, UBL4A
Abstract
BCL2-associated athanogene 6 (BAG6) is a member of the BAG protein family, which is implicated in diverse cellular processes including apoptosis, co-chaperone, and DNA damage response (DDR). Recently, it has been shown that BAG6 forms a stable complex with UBL4A and GET4 and functions in membrane protein targeting and protein quality control. The BAG6 sequence contains a canonical nuclear localization signal and is localized predominantly in the nucleus. However, GET4 and UBL4A are found mainly in cytoplasm. Whether GET4 and UBL4A are also involved in DDR in the context of the BAG6 complex remains unknown. Here, we provide evidence that nuclear BAG6-UBL4A-GET4 complex mediates DDR signaling and damage-induced cell death. BAG6 appears to be the central component for the process, as depletion of BAG6 leads to the loss of both UBL4A and GET4 proteins and resistance to cell killing by DNA-damaging agents. In addition, nuclear localization of BAG6 and phosphorylation of BAG6 by ATM/ATR are also required for cell killing. UBL4A and GET4 translocate to the nucleus upon DNA damage and appear to play redundant roles in cell killing, as depletion of either one has no effect but co-depletion leads to resistance. All three components of the BAG6 complex are required for optimal DDR signaling, as BAG6, and to a lesser extent, GET4 and UBL4A, regulate the recruitment of BRCA1 to sites of DNA damage. Together our results suggest that the nuclear BAG6 complex is an effector in DNA damage response pathway and its phosphorylation and nuclear localization are important determinants for its function.
Introduction
DNA damage response (DDR)4 is an evolutionally conserved pathway that coordinates cell cycle check point activation, DNA repair, and cellular apoptosis, which is essential for the maintenance of genomic integrity (1). Central to DDR are two kinases, ATM and ATR, which are activated primarily through DNA double-strand breaks and replication stress, respectively, and initiate the signaling transduction cascade by phosphorylating their substrates. ATM and ATR are members of the phosphatidylinositol 3-kinase-related kinase family that preferentially phosphorylate substrates at serine or threonine residue followed by a glutamine (S/TQ motif). Genome-wide screens for ATM/ATR substrates using phospho-specific S/TQ antibodies have identified >700 potential ATM/ATR substrates, revealing extensive signaling networks involved in the rapid and complex response to DNA damage (2, 3).
One of the putative ATM/ATR substrates identified in our screen is BAG6 (also known as BAT3/Scythe) (3). The BAG6 gene, located on chromosome 6 within the class III region of human major histocompatibility complex, encodes the 124-kDa BAG6 protein (4), a member of BAG family proteins involved in diverse biological processes (2). Initially identified in Xenopus as a binding partner of a potent apoptosis inducer Reaper, BAG6/Scythe was shown as a positive regulator of apoptosis (5, 6). Depletion of BAG6 prevents Reaper-induced apoptosis, and expression of an N-terminal domain truncated BAG6 alone is able to induce apoptosis in vitro (5). Moreover, BAG6 also plays an important role in DNA damage-induced and endoplasmic reticulum stress-induced apoptosis (7, 8). Recently, BAG6/BAT3 has been shown to facilitate DOT1L-mediated H3K79 dimethylation and promote DNA damage-induced 53BP1 foci formation at G1/G2 cell cycle phases (9). Consistent with the notion that BAG6 is involved in diverse cellular processes, including cell cycle progression, T cell function, immune response, and differentiation, targeted deletion of Bag6 in mice results in lethality associated with pronounced developmental defects in the lung, kidney, and brain (10). Importantly, many of the BAG6 functions are operated through Hsp70-mediated protein refolding and ubiquitin-mediated proteolysis of misfolded proteins (11–13).
Several recent studies reported the identification of a stable ternary complex formed by BAG6 with GET4 (also known as C7orf20 and Trc35) and UBL4A (also known as Get5 and Gdx), two well defined components of the tail-anchored protein targeting pathway, which facilitates the delivery of tail-anchored proteins to endoplasmic reticulum (ER) (13, 14). Moreover, the BAG6 complex also functions as a regulator in the endoplasmic reticulum-associated degradation (ERAD) system (15). BAG6 exhibits in vitro holdase activity that maintains the unfolded polypeptides in soluble state thereby enhancing the efficiency of ERAD. Because both tail-anchored targeting and ERAD take place in the cytoplasm, together these studies suggest that BAG6 complex regulates cellular protein quality control in cytoplasm by targeting multiple substrates for a variety of cellular functions.
The BAG6 sequence contains a canonical bipartite nuclear localization signal (NLS) near its C terminus. As expected, immune-fluorescence staining of exogenously expressed BAG6 shows a predominantly nuclear localization, and mutating the canonical NLS resulted in BAG6 accumulation in cytoplasm (16). Endogenous BAG6 is found in both nucleus and cytoplasm and is shuttled between the two compartments in a cell cycle-dependent manner (17). Interestingly, both exogenously expressed GET4 and UBL4A localize predominantly in the cytoplasm, and overexpressed GET4 is able to retain both endogenous and exogenously expressed BAG6 in the cytoplasm (15). Therefore, questions remain as to whether UBL4A and GET4 in the context of the BAG6 complex also play a role in DDR, a process that involves predominantly nuclear proteins. Here, we provide evidence that the BAG6 complex is a component of the DDR signaling network. We show that nuclear BAG6 is a substrate of ATM/ATR kinase and that BAG6 nuclear localization and phosphorylation are essential for cellular sensitivity to DNA damage. Importantly, UBL4A and GET4, which localize mainly in the cytoplasm in unstressed cells, translocate to nuclear upon DNA damage, and together the BAG6 complex is required for proper recruitment of DDR factors to sites of DNA damage.
EXPERIMENTAL PROCEDURES
Cell Culture, Chemicals, and Antibodies
HeLa, MCF7, FT169 (A-T cells), and YZ5 (ATM-complemented A-T cells) were maintained in DMEM, and U2OS cells in McCoy's 5A medium, all supplemented with 10% fetal bovine serum (FBS). The stable ATM knockdown cell line was a generous gift from Dr. B. Chen (University of Texas Southwestern). Doxorubicin, chloroquine, MG132, lactacystin, and ATM inhibitor KU55933 were purchased from Sigma.
Antibodies used in the study are as follows: anti-BAG6 and ATM from Abcam; anti-C7ORF20/GET4, Brca1-pS1457, ATM-pS1981, Brca1-pS1387, SMC1-pS966, ATR, and V5 from Bethyl Laboratories; anti-UBL4A, β-tubulin, and FLAG (M2) from Sigma; Myc (9E10) and GFP from Santa Cruz Biotechnology. For Western blotting, all antibodies were diluted 1:1000, except for BAG6, which was diluted 1:10,000.
Expression Vectors and Stable Cell Lines
Full-length GFP-BAG6 was a generous gift from Dr. S. Kornbluth (Duke University). Human GET4 and UBL4A were cloned into pcDNA4/TO/myc-HisB (Invitrogen) and pEYFP-C1 (Invitrogen), respectively, from a cDNA pool of HeLa cells. V5-BAG6-WT was generated by subcloning BAG6 to pcDNA3.1-V5 (inserted at BamHI/EcoRI sites) vector. Phosphorylation mutant (V5-BAG6-S212A) was generated by BAG6-Ser212 substitution to alanine. Nuclear localization mutant (V5-BAG6-NLSmt) was made as described in Ref. 16. DNA transfections were performed using TransIT-LT1 reagent (Mirus) or Lipofectamine 2000 (Invitrogen). To generate GFP-BAG6-WT, V5-BAG6-WT, V5-BAG6-S212A, V5-BAG6-NLSmt expressing cell lines, U2OS cells were transfected with the DNA plasmids using Lipofectamine 2000 reagent, and stable clones were selected with Geneticin (GE Healthcare). Single clones expressing comparable amounts of exogenous proteins were used in the experiments.
RNA Interference
siRNA used in this work were synthesized by IDT, Sigma, or Invitrogen. SiRNA transfections were carried out using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocols. The siRNA sequences for ATR and BAG6 were described in Refs. 18 and 7, respectively. siRNA against the BAG6–3′-UTR region is GCUCUAUGGCCCUUCCUCA; control siRNAs (NC1 or NC2) were purchased from Sigma. The siRNA sequences for UBL4A and GET4 are: UBL4A-1, CCCUGGAGAAGGUGCUACUAGAAGA; UBL4A-2, UCCAAAGUCUUGGCCCGCCACUUCA; UBL4A-3′-UTR, 5′-GGCACAAGAGGACAUCUAACCACCT-3′ and 3′-ACCCGUGUUCUCCUGUAGAUUGGUGGA-5′; GET4–1, GAGAUUGACCCACAAUAAA; GET42, GUGCUACAGUUUCUCUGUU.
Immunofluorescence
Cells grown on poly-d-lysine-coated coverslips were fixed with 4% formaldehyde (w/v) and 2% sucrose in PBS and permeabilized in 0.5% Triton X-100 in PBS. Cells were incubated with primary antibody in 2% BSA/PBST for 1 h followed by Alexa Fluor 488-or 594-conjugated secondary antibodies for 30 min and counterstained with DAPI. Images were captured on a Nikon Eclipse TE2000-U fluorescence microscope controlled by NIS-Elements (Nikon).
Cell Survival and Chromatin Fractionation Assays
Cell survival was determined with WST-1 assay (Roche Applied Science). Chromatin fractionations were performed as described in Ref. 19. Briefly, cells were harvested by trypsinization, washed with PBS, and resuspended in buffer A (10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 0.34 m sucrose, 10% glycerol, 1 mm DTT, and protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and cells were incubated for 5 min on ice. Cytosolic proteins (S1) were separated from the nuclei by low speed centrifugation (1300 × g for 4 min at 4 °C). Isolated nuclei were washed once with buffer A and lysed in buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, protease inhibitors). After a 10-min incubation on ice, soluble nuclear proteins (S2) were separated from chromatin by centrifugation (1700 × g for 4 min at 4 °C). Finally, chromatin (P3) was resuspended in Laemmli buffer and sheared by sonication.
Quantification and Statistical Analysis
ImageJ (National Institutes of Health, Bethesda, MD) was used for Western blot quantification. Protein levels were normalized to loading controls unless indicated otherwise. Statistical analyses were performed using Student's two-tailed t test.
RESULTS
GET4 and UBL4A Are Associated with BAG6
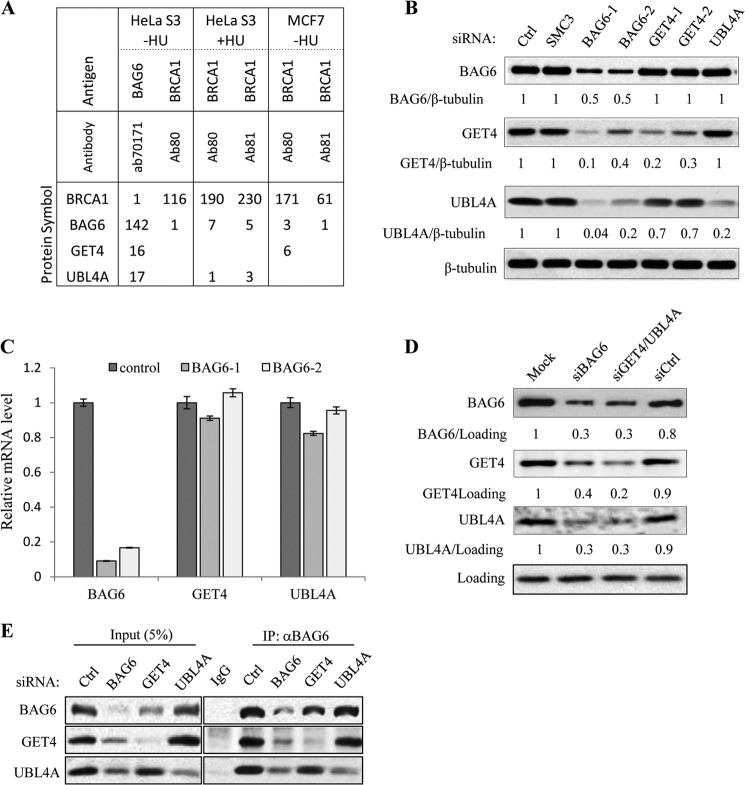
We originally identified BAG6 as a putative ATM/ATR substrate through its cross-reactivity to multiple pSQ antibodies (3). To analyze the signaling pathways that BAG6 functions, we isolated endogenous nuclear BAG6 complexes from cycling and hydroxyurea-treated HeLa and MCF7 cells and identified the associated proteins by mass spectrometry (MS). Consistent with previously published results (13–15), we repeatedly detected UBL4A and GET4 in BAG6 IP (Fig. 1, A and E). We also detected small number of BRCA1 peptides from BAG6 IP and components of the BAG6 complex in BRCA1 IP. To determine the relationship among the three components, we knocked down each protein with siRNA and examined how the complex formation was affected. Interestingly, when BAG6 was knocked down, levels of UBL4A and GET4 were greatly reduced (Fig. 1B); in contrast, the BAG6 level was not affected by either GET4 or UBL4A knockdown. Knocking down GET4 also resulted in a slight reduction of UBL4A protein level, whereas knocking down UBL4A had no effect on GET4. However, when both GET4 and UBL4A were knocked down, the BAG6 protein level was reduced by >3-fold (Fig. 1D). Real-time quantitative PCR analysis showed that mRNA levels of UBL4A and GET4 remained at levels similar to those in control knockdown (Fig. 1C), suggesting that BAG6 is required to maintain GET4 and UBL4A at the protein level. However, neither GET4 nor UBL4A protein can be restored by treating the cells with proteasome or lysosome inhibitors (supplemental Fig. 1, A and B). Next, we investigated whether GET4 or UBL4A is required for the BAG6 complex formation. To this end, we performed IP with a BAG6 antibody in HeLa cells that were transfected with either GET4 or UBL4A siRNA. As shown in Fig. 1E, loss of UBL4A had no effect on the amount of GET4 associated with BAG6, and loss of GET4 had no effect on the amount of UBL4A associated with BAG6. Together these data suggest that BAG6 plays an organizing role in the complex to maintain the integrity of the complex and that GET4 and UBL4A can independently interact with BAG6.
FIGURE 1.
UBL4A and GET4 are components of the BAG6 complex. A, number of peptide-spectrum matches of BRCA1 and BAG6 complex components identified by mass spectrometry. Immunoprecipitation was performed with nuclear extracts prepared from HeLa S3 suspension or MCF7 cells not treated or treated with hydroxyurea (HU). B, protein levels of BAG6, GET4, and UBL4A in HeLa cells transfected with the indicated siRNAs determined by Western blotting. Control (Ctrl) and SMC3 siRNA transfections served as negative controls. β-Tubulin served as a loading control. C, knockdown of BAG6 gene performed as described in B. mRNA levels of BAG6, UBL4A, and GET4 were assessed by Real-time quantitative PCR. Relative mRNA expression was normalized to 34B4 transcript. D, HeLa cells mock transfected or transfected with nontargeting siRNA (siCtrl) or siRNA against BAG6, GET4, and UBL4A together. Whole cell extracts were analyzed by Western blotting E, immunoprecipitation of endogenous BAG6 performed with whole cell lystes prepared from HeLa cells depleted of BAG6, GET4, and UBL4A.
GET4 and UBL4A Translocate from Cytoplasm to Nucleus upon DNA Damage in a BAG6-dependent Manner
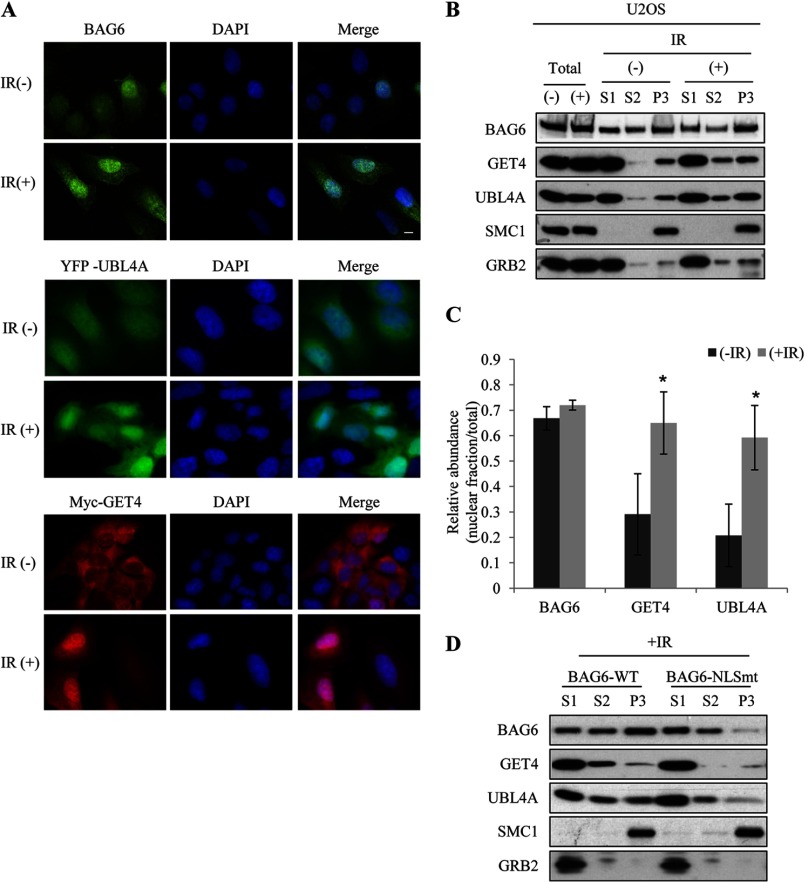
Previously published results suggest that the BAG6 complex plays an important role in protein quality control in several cellular processes that take place in the cytoplasm. However, BAG6 contains a canonical NLS and was found in both nucleus and cytoplasm (16). We next examined the subcellular localization of BAG6 complex in cycling and IR-treated cells by immunostaining. Consistent with previous reports, BAG6 exhibited strong staining in the nucleus in normal cycling cells and remained in the nucleus following ionizing irradiation (Fig. 2A). Because no commercial antibodies that specifically recognize endogenous GET4 and UBL4A by immunofluorescence are available, we generated U2OS cell lines stably expressing Myc-GET4 and YFP-UBL4A. Consistent with published results (15) Myc-GET4 was found predominantly in the cytoplasm, whereas YFP-UBL4A was found in both cytoplasm and nucleus (Fig. 2A). After IR, both GET4 and UBL4A accumulated in the nucleus, indicating their translocation to nucleus. To confirm the immunostaining results and obtain independent evidence for endogenous GET4 and UBL4A translocation, we performed chromatin fractionation in HeLa and U2OS cells and detected the protein distribution by Western blotting. As shown in Fig. 2, B and C, and supplemental Fig. 2, both UBL4A and GET4 were localized predominantly in the cytoplasm (S1) in normal cycling cells, but were enriched in soluble nuclear fraction (S2) and chromatin-bound fractions (P3) upon DNA damage; in contrast, BAG6 was distributed in all three fractions before IR, and DNA damage did not cause significant change in its localization (Fig. 2C). To determine whether DNA damage-induced GET4 and UBL4A translocation depends on BAG6, we performed chromatin fractionation experiments in U2OS cells stably expressing BAG6 nuclear localization signal mutant (NLSmt). As shown in Fig. 2D, DNA damage-induced nuclear enrichment of GET4 and UBL4A were reduced in NLSmt-expressing cells, indicating that GET4 and UBL4A translocation depends on nuclear BAG6.
FIGURE 2.
UBL4A and GET4 translocate to nucleus upon DNA damage. A, U2OS cells or U2OS cells stably expressing Myc-GET4 or YFP-UBL4A were treated with IR. Subcellular localizations of indicated proteins were examined by immunostaining (anti-BAG6 and Myc) or directly with YFP fluorescence signals. B, chromatin fractionations were performed using untreated or IR-treated U2OS cells. S1, cytoplasm fraction; S2, soluble nucleus fraction; and P3, chromatin-bound fraction. SMC1 and GRB2 serve as fractionation control for nuclear and cytoplasmic proteins, respectively. C, quantification of nuclear fraction (S2+P3) of BAG6 complex was done before and after IR. Western blotting signals of nuclear fraction were normalized to total (S1+S2+P3) protein levels. All error bars represent mean ± S.D. (n = 3). *, p < 0.05. D, chromatin fractionation was performed on IR-treated U2OS cells stably expressing wild-type BAG6 (BAG6-WT) or BAG6 nuclear localization mutant (BAG6-NLSmt).
BAG6 Is Phosphorylated in Response to DNA Damage in an ATM/ATR-dependent Manner
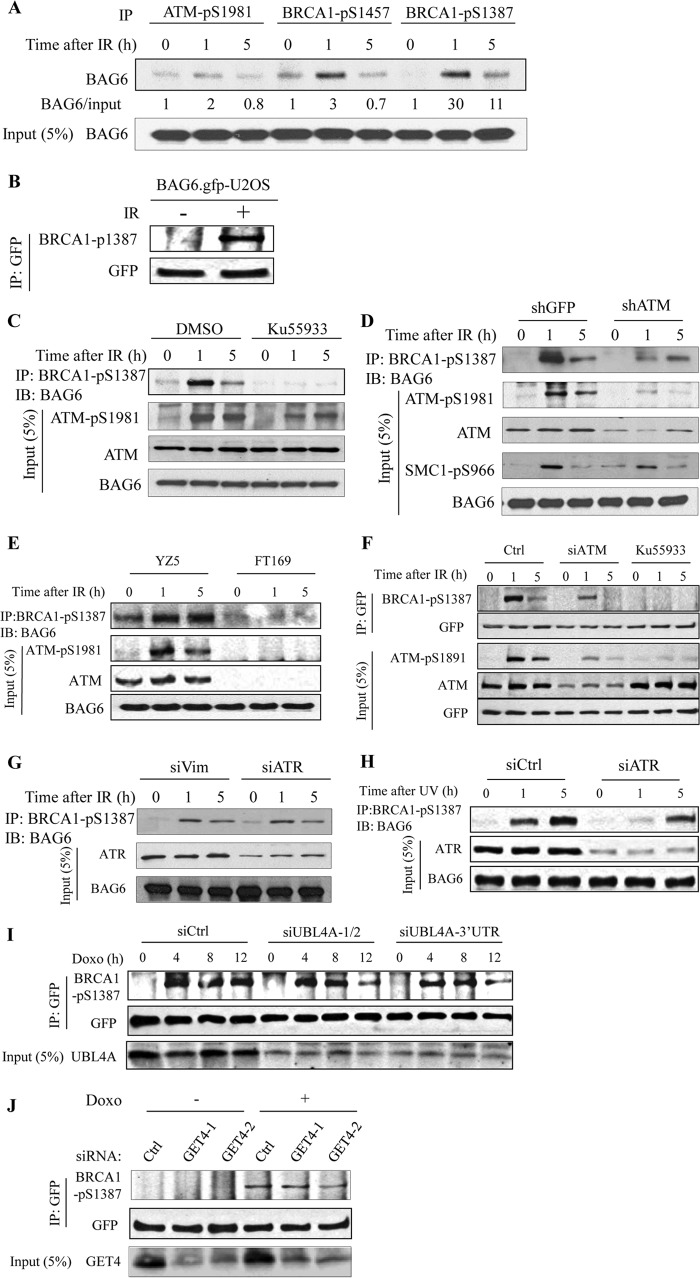
BAG6 was previously identified as a putative substrate of ATM/ATR kinase, as it cross-reacts to several pS/TQ antibodies including anti-BRCA1-pS1457, anti-BRCA1-pS1387, and anti-ATM-pS1981 antibodies (3). To confirm that BAG6 is a substrate of ATM/ATR, we immunoprecipitated the phospho-proteins from IR-treated HeLa cells with each of these antibodies and immunoblotted them with an anti-BAG6 antibody. As shown Fig. 3A, all three antibodies were able to recognize phosphorylated BAG6 from cells exposed to DNA damage, and BRCA1-pS1387 showed the best induction. We also immunoprecipitated GFP-tagged BAG6 from GFP-BAG6 stably overexpressing U2OS cells with a GFP antibody and detected phosphorylated exogenous BAG6 with BRCA1-pS1387 antibody (Fig. 2B). These results suggest that BAG6 is indeed phosphorylated and is unlikely co-precipitated with other phosphorylated proteins.
FIGURE 3.
BAG6 is phosphorylated in an ATM/ATR-dependent manner. A, HeLa cells treated with IR (10 Gy) were subjected to immunoprecipitation with the indicated phospho-antibodies. Phosphorylated BAG6 was detected with BAG6 antibody. B, U2OS cells stably expressing GFP-BAG6 were treated with IR (10 Gy) and subjected to GFP IP. Phosphorylation of GFP-BAG6 was detected by Western blotting (IB) with anti-BRCA1-pS1387 antibody. C, HeLa cells were pretreated with an ATM kinase inhibitor KU55933 for 1 h before IR (10 Gy). Cells were harvested at indicated times after exposure to IR. Phosphorylated BAG6 was immunoprecipitated with anti-BRCA1-pS1387 antibody and detected with a BAG6 antibody. D, HeLa cells stably knocked down ATM (shATM) or control (shGFP) were treated with 10 Gy of IR and harvested at the indicated times. Phosphorylated BAG6 was immunoprecipitated with anti-BRCA1-phospho-S1387 antibody and detected by Western blotting with a BAG6 antibody. E, YZ5 (A-T +ATM) and FT169 (A-T) cells were treated with IR (10 Gy) and harvested at indicated times. Phosphorylated BAG6 was immunoprecipitated with anti-BRCA1-pS1387 antibody and detected by Western blotting with a BAG6 antibody. F, U2OS cells stably expressing GFP-BAG6 were either transfected with the indicated siRNAs or pretreated with an ATM kinase inhibitor KU55933 for 1 h before IR. Phosphorylation of GFP-BAG6 was detected as described in B. G and H, HeLa cells were transfected with the indicated siRNAs and were treated with 10 Gy of IR or UV (60 Joules/m2). BAG6 phosphorylation was assessed as in C. I and J, U2OS cells stably expressing GFP-BAG6 were transfected with indicated siRNAs and were treated with doxorubicin (Doxo; 2 μg/ml). Phosphorylation of GFP-BAG6 was assessed as in B.
To test whether BAG6 is an ATM substrate, we inhibited ATM kinase activity by pretreating the HeLa cells with an ATM-specific inhibitor KU55933 and examined BAG6 phosphorylation by BRCA1-pS1387 IP followed by BAG6 Western blotting. Successful inhibition of ATM kinase activity was confirmed by the reduction of ATM autophosphorylation (ATM-pS1981) (Fig. 3C). A decrease in BAG6 phosphorylation, but not a decrease in total BAG6 protein level, was observed in the IR-treated cells, indicating that BAG6 phosphorylation depends on ATM activity. Consistent with these data, BAG6 phosphorylation was also compromised in a fibroblast cell line derived from A-T patient (FT169) and an ATM stable knockdown cell line as well as siATM-transfected GFP-BAG6 stable cells (Fig. 3, D–F). ATM/ATR is another kinase that shares substrate consensus similar to that of ATM, but is activated mainly in response to DNA replication block and to IR at later times (20). Because both ATM and ATR may phosphorylate BAG6, we tested whether ATR is also involved in BAG6 phosphorylation. We transiently knocked down ATR by siRNA and found that depletion of ATR did not affect BAG6 phosphorylation upon IR treatment (Fig. 3G), suggesting that ATR is not the major kinase for IR-induced phosphorylation; in contrast, phosphorylation of BAG6 was compromised upon UV treatment (Fig. 3H), Together, these data suggest that BAG6 is phosphorylated upon DNA damage.
Next, we asked whether GET4 and UBL4A regulate BAG6 phosphorylation. To this end, we transiently knocked down GET4 and UBL4A in U2OS cells stably expressing GFP-BAG6 with multiple siRNA sequences and examined BAG6 phosphorylation upon doxorubicin treatment, which also generates double-stranded DNA breaks. We immunoprecipitated GFP-BAG6 and detected its phosphorylation by BRCA1-pS1387 Western blotting. As shown in Fig. 3I, knocking down UBL4A resulted in a marked reduction of BAG6 phosphorylation compared with a control knockdown at 12 h after doxorubicin treatment; in contrast, knockdown GET4 had no obvious effect on BAG6 phosphorylation (Fig. 3J). Taken together, our data suggest that BAG6 is phosphorylated upon DNA damage in an ATM-dependent manner and that UBL4A is required for maintaining the BAG6 phosphorylation.
BAG6 Phosphorylation at Serine 212 in the Nucleus Is Necessary for DNA Damage Sensitivity
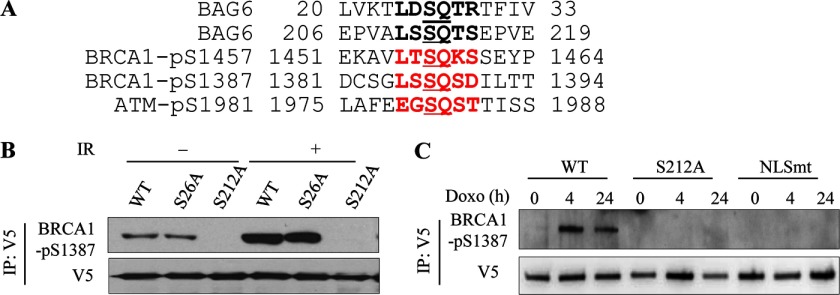
BAG6 protein has five potential ATM/ATR phosphorylation S/TQ consensus motifs. Comparison of the BAG6 sequence with the phospho-antibody recognition motifs predicts Ser26 and Ser212 as the most possible phosphorylation sites (Fig. 4A). We therefore mutated these two serine residues to alanines (S26A or S212A) and tested DNA damage-induced BAG6 phosphorylation. We used U2OS cells stably overexpressing V5-tagged wild-type (WT), S26A, or S212A BAG6 mutants and examined phosphorylated BAG6 protein by V5 IP followed by anti-BRCA1-pS1387 immunoblotting. As shown in Fig. 4B, whereas IR-induced phosphorylation of BAG6-S26A was not affected, phosphorylation of BAG6-S212A was almost completely abolished, suggesting that Ser212 is the major BAG6 phosphorylation site. In addition, BAG6 phosphorylation in a mutant that localizes mainly in the cytoplasm (NLSmt) was also abolished, suggesting that the phosphorylation occurs in the nucleus (Fig. 4C).
FIGURE 4.
BAG6 protein is phosphorylated at Ser212 in response to DNA damage. A, alignment of BRCA1 and ATM phosphorylation motifs and potential BAG6 phosphorylation motifs. B, U2OS cells stably overexpressing V5-tagged wild-type (WT) or BAG6 point mutants (S26A, S212A) immunoprecipitated with anti-V5 antibody. BAG6 phosphorylation was assessed by Western blotting using anti-BRCA1-pS1387 antibody. C, U2OS cells stably expressing V5 tagged BAG6 and treated with doxorubicin (Doxo; 2 μg/ml) for the indicated times. Phosphorylation of V5-BAG6-WT was assessed by immunoprecipitation with anti-V5 antibody and immunoblotting with BRCA1-pS1387 antibody.
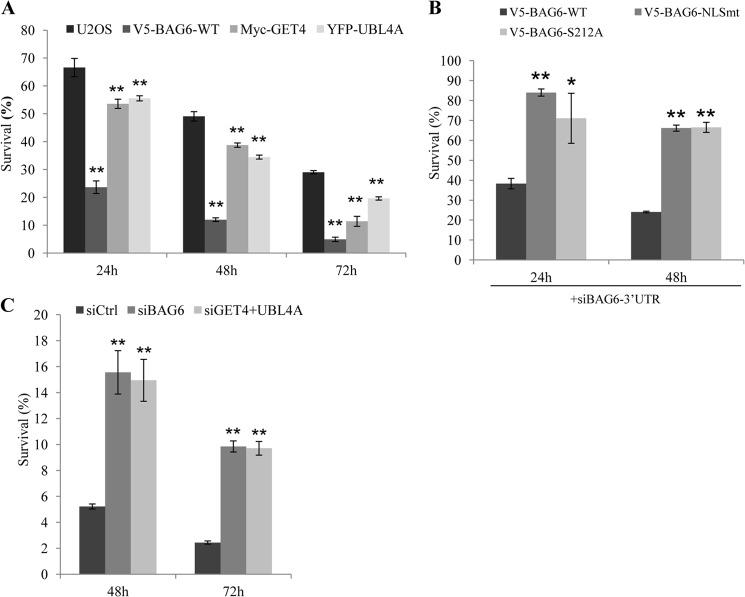
Initially identified as apoptosis regulator, BAG6 has been shown to mediate apoptosis induced by several agents, including ER stress (8) and DNA damage (7). To test whether GET4 and UBL4A also mediate this function, we treated U2OS stably expressing V5-BAG6, Myc-GET4, and YFP-UBL4A with doxorubicin and measured cell viability using the WST-1 assay. Consistent with published data, we found that cells overexpressing WT-BAG6 were significantly more sensitive to doxorubicin-induced cell death than parental U2OS cells (Fig. 5A). Overexpressing Myc-GET4 or YFP-UBL4A resulted in a moderate increase of doxorubicin-induced cell death (Fig. 5A), suggesting that they play less important roles than BAG6 in cell death. To test whether BAG6 phosphorylation is required for this function, we performed the similar experiments with U2OS cells stably expressing S212A and NLS mutants of BAG6. As shown in supplemental Fig. 3A, cells overexpressing BAG6 NLS mutant or phosphorylation S212A mutant are less sensitive to doxorubicin-induced cell death than that of U2OS cells, suggesting that these mutants failed to potentiate DNA damage-induced cell death. Similar results were also obtained when the endogenous BAG6 was knocked down by 3′-UTR targeting siRNA (Fig. 5B). Next, we investigated DNA damage sensitivities in U2OS cells depleted of BAG6, GET4, and UBL4A and found that whereas loss of GET4 or UBL4A alone had no significant effect on survival upon DNA damage (supplemental Fig. 3B), knocking down BAG6 or simultaneously knocking down both GET4 and UBL4A resulted in moderate resistance to DNA damage (Fig. 5C). As the BAG6 protein level is reduced in GET4 and UBL4A double knockdown, together our data suggest that a reduction of BAG6 is correlated with DNA damage resistance.
FIGURE 5.
BAG6 nuclear localization and phosphorylation potentiate DNA damage induced cell death. A, U2OS cells and U2OS cells stably expressing the indicated proteins were treated with doxorubicin (2 μg/ml). Cell survival was determined using WST-1 assay at indicated times. B, U2OS cells stably expressing wild-type BAG6 (WT), BAG6 nuclear localization mutant (NLSmt), BAG6-S212A phospho-mutant (S212A) were transfected with an siRNA targeting the 3′-UTR region of BAG6 to knock down endogenous BAG6. Cells were treated with doxorubicin and assayed for survival as described in A. C, HeLa cells transfected with indicated siRNAs were treated with doxorubicin and assayed for cell survival at indicated times. All error bars represent mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01.
BAG6 Complex Regulates IR-induced BRCA1 Focus Formation
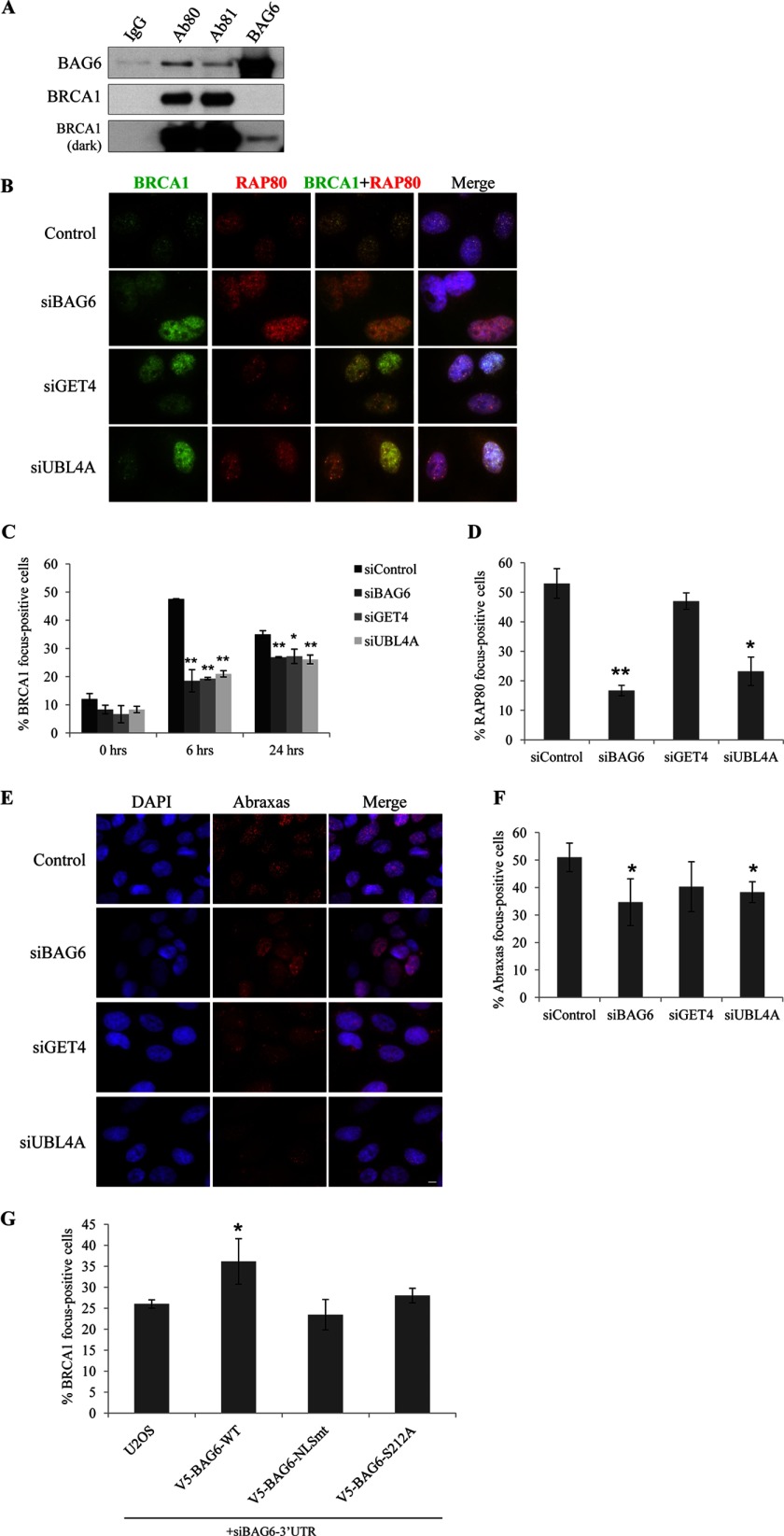
BRCA1, the familial breast tumor suppressor, plays an important role in maintaining genome integrity. Upon exposure to DNA damage, BRCA1 translocates to sites of the damage, where it mediates DDR signaling and homologous recombination-mediated repair. We detected by mass spectrometry a small number of BRCA1 peptides in BAG6 IP and BAG6 components in BRCA1 IP (Fig. 1A). We confirmed the interaction with BAG6 and BRCA1 IP followed by Western blotting (Fig. 6A). To determine whether BAG6 complex participates in BRCA1-mediated response, we examined IR-induced BRCA1 focus formation in cells depleted each component of the BAG6 complex. As shown in Fig. 6, C and D, BAG6, GET4, and UBL4A knockdown cells exhibited a marked reduction of IR-induced BRCA1 foci. In addition, focus formation of RAP80 and Abraxas was also impaired in BAG6 and UBL4A knockdown cells, but not in GET4 knockdown cells (Fig. 6, E–G). Knocking down endogenous BAG6 with 3′-UTR targeting siRNA in BAG6 stable cells showed that exogenous GFP-BAG6 can partially restore DNA damage sensitivity (Fig. 3G). Finally, we examined IR-induced γH2AX in the absence of BAG6 complex (supplemental Fig. 4) and found a moderate reduction of γH2AX focus-forming cells at 6 h after IR, but not at later times. These results suggest that double-strand break repair function of BAG6 knockdown cells remains largely intact.
FIGURE 6.
BAG6 complex regulates IR-induced BRCA1 focus formation. A, co-immunoprecipitation of endogenous BAG6 and BRCA1 from HeLa cell nuclear extract followed by Western blotting. Two BRCA1 antibodies (Ab80 and Ab81) and BAG6 antibody were used for IP. B, representative images of DNA damage-induced BRCA1 and RAP80 foci in U2OS cells depleted of BAG6 complex.U2OS cells were transfected with the indicated siRNA and treated with 10 Gy IR. Cells were fixed at 8 h after IR and stained with indicated antibodies. The scale bar represents 10 μm. C and D, quantification of BRCA1 and the RAP80 foci containing U2OS cells depleted of BAG6 complex. Cells with >15 foci were counted as focus-positive cells. At least 300 cells were counted for each time point. All error bars represent mean ± S.D. (n = 3). *, p < 0.05; **, p < 0.01. E, representative images of Abraxas immunostaining of U2OS cells transfected with the indicated siRNAs. G, U2OS or U2OS stably expressing indicated BAG6 proteins and transfected with a siRNA targeting 3′-UTR of BAG6. Cells forming IR-induced foci were counted as described above.
DISCUSSION
BAG6 Plays a Major Role in Complex Organization
We show that, among the three components, BAG6 plays a more important role in the complex organization, as loss of BAG6 results in the reduction of GET4 and UBL4A proteins, whereas loss of either GET4 or UBL4A alone has no obvious effect on BAG6 level; however, loss of GET4 and UBL4A together leads to the reduction of BAG6 protein Because mRNA levels of the two proteins were not affected, it suggests that the regulation is at protein level.
Interestingly, whereas GET4 and UBL4A (Get5 in yeast) are evolutionarily conserved from yeast to human, BAG6 is only found in higher eukaryotes. Yeast Get4 and Get5 form a stable complex (Get4/5) without Bag6, but Get5 is considerably larger in size than its human counterpart UBL4A, containing an N-terminal domain (Get5-N) that is not found in higher eukaryotes (7). Structural studies of Get4/5 complex revealed that the Get5 N-terminal domain mediates its interaction with Get4, whereas the Get5 C terminus mediates Get4/5 dimerization (9, 21). However, as the higher eukaryotes do not have the Get5-N domain, it was speculated that another protein that contains a Get5-N-like region could substitute this domain for Get4 binding (21). Our results suggest that BAG6 can indeed functionally substitute the Get5-N domain for the stabilization of GET4 and UBL4A; however, structural analysis is necessary to provide direct evidence.
BAG6 Complex Is an Integral Component of the DNA Damage Response Network
It has been shown previously that BAG6 protein plays an important role in DNA damage-induced cell death (21) and the formation of IR-induced 53BP1 foci (22). However, it is not clear whether GET4 and UBL4A, which mainly localize in the cytoplasm, also participate in DDR. Here we present evidence that nuclear BAG6 complex containing both UBL4A and GET4 is an integral component of the DNA damage response network. We show that BAG6 is phosphorylated in an ATM/ATR-dependent manner at Ser212, an S/TQ consensus motif, and that maintaining this phosphorylation depends on one of its partner UBL4A; further we show that BAG6 phosphorylation in the nucleus is necessary for this function. Importantly, GET4 and UBL4A, which are primarily located in the cytoplasm, translocate to the nucleus upon DNA damage in a BAG6-dependent manner. Moreover, depletion of BAG6 compromises the formation of DNA damage-induced BRCA1 foci and leads to resistance to DNA damage-induced cell death.
Our finding that depletion of BAG6 also results in simultaneous loss of GET4 and UBL4A raises an important question regarding all functional studies of BAG6, namely, whether the cellular phenotypes observed in BAG6 knockdown or knock-out are attributed to loss of BAG6 itself or to that of all three components in the BAG6 complex. Our data suggest that whereas some DDR defects arise from loss of each individual component, others are attributed mainly to loss of BAG6. We show that overexpression of BAG6 can potentiate DNA damage-induced cell death, and siRNA-mediated knockdown of BAG6 leads to resistance; however, knockdown of GET4 or UBL4A alone has negligible effect on cell death. Importantly, when both GET4 and UBL4A are knocked down simultaneously, which results in a reduction of BAG6, cells are more resistant to killing by DNA-damaging agents, indicating that the effect is likely caused by loss of BAG6 protein. It appears that whereas the intact complex is necessary for timely DDR signaling, GET4 and UBL4A are dispensable for induction for cell death. Follow-up investigations to identify the direct targets in each pathway will provide insights at molecular levels.
There also appears to be a subtle difference between GET4 and UBL4A in their involvement in DDR: whereas knocking down GET4 does not affect BAG6 phosphorylation, RAP80 and Abraxas recruitment, knocking down UBL4A results in weakened BAG6 phosphorylation at later times in response to IR and a moderate reduction of RAP80 and Abraxas recruitment. There results suggest that UBL4A may play a more direct role in DDR. Interestingly, the role of UBL4A in ERAD also appears to be different from that of GET4/Trc35 (15). Although UBL4A is required for efficient degradation of misfolded ER proteins, it is dispensable for the solubility of ERAD substrates. On the other hand, GET4/Trc35 may play an essential role in maintaining a cytosolic pool of BAG6, as well as in maintaining the solubility of ERAD substrates.
Does BAG6 Complex Control Multiple Cellular Processes through Modulating Protein Ubiquitination?
BAG6 plays an essential role in multiple cellular processes. Thus far, a common underlying theme of most of the functions is associated with its co-chaperone activities. The presence of the UBL domains in BAG6 and UBL4A provides a tantalizing clue, linking the BAG6 complex to protein ubiquitination system. From this perspective, it is not surprising to uncover a role of the BAG6 complex in DDR, as protein ubiquitination has emerged as an important regulatory mechanism for DDR signaling (1, 23). An expanding list of E3 ligases and ubiquitin hydrolases (RNF8, RNF168, KAP1/Trim28, BRCA1, BARD1, USP7, UP1, and BRCC36, to name a few) have been reported to function at multiple stages in DDR including damage detection, signal amplification, and termination. It is conceivable that protein quality control plays a crucial role to ensure proper response when tens, perhaps hundreds of proteins rapidly translocate to sites of damage, and is particularly important for large proteins such as BRCA1. The identification of direct targets of the BAG6 complex in DDR signaling and cell death holds the key to the understanding of its functions in DDR at molecular levels.
A Model for BAG6 Function in DDR
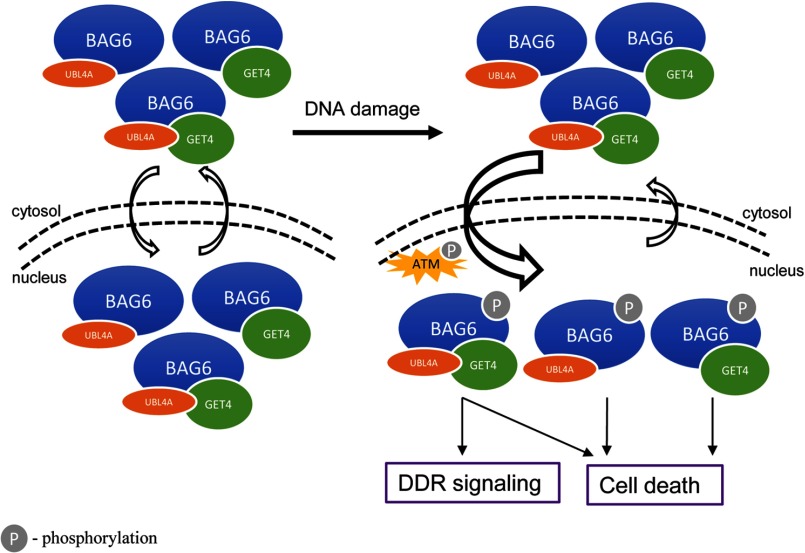
Based on current and published data, we propose a highly speculative model to explain how three components of the BAG6 complex play differential roles in DDR signaling and cell death (Fig. 7). It appears that BAG6 can exist in at least three different complexes containing BAG6-UBL4A, BAG6-GET4, and BAG6-UBL4A-GET4 (supplemental Fig. 5), i.e. it needs at least one partner to be stabilized. This arrangement is consistent with our data that GET4 and UBL4A can interact with BAG6 independently of the presence of the other. These BAG6 complexes are shuttled between cytoplasm and nucleus possibly in a cell cycle-dependent manner (17). Upon DNA damage, checkpoint activation leads to ATM/ATR-dependent BAG6 phosphorylation and either increased nuclear import, or decreased nuclear export, resulting a net accumulation of nuclear complexes. Although the tertiary complex is required for timely assembly of DDR signaling complex at sites of damage, all three complexes are functional in the induction of cell death too. This functional redundancy is consistent with our data that knockdown either GET4 or UBL4A does not appreciably affect cell death, but simultaneous knockdown of both results in resistance. It would also predict that the direct target(s) in the cell killing pathway binds to BAG6, regardless of its partner(s). Finally, because co-IP experiment cannot distinguish the three BAG6 complexes, structural study is the ultimate tool to investigate how BAG6 complexes are organized.
FIGURE 7.
Working model for BAG6 complex function in DDR.
Supplementary Material
Acknowledgments
We thank Drs. Sally Kornbluth (Duke University), Benjamin Chen (University of Texas Southwestern), Bin Wang (M. D. Anderson Cancer Center), and Junjie Chen (M. D. Anderson Cancer Center) for providing reagents; and Joel M. Sederstrom for expert assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM080703 and Chinese Natural Science Foundation Grant 31270822. This work was also supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the National Institutes of Health Grants AI036211, CA125123, and RR024574.

This article contains supplemental Figs. 1–5.
- DDR
- DNA damage response
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3-related protein
- BRCA1
- breast cancer type 1 susceptibility protein
- ER
- endoplasmic reticulum
- ERAD
- endoplasmic reticulum-associated degradation
- GET4
- Golgi to ER traffic protein 4
- Gy
- gray
- IP
- immunoprecipitation
- IR
- ionizing radiation
- NLS
- nuclear localization signal
- RAP80
- receptor-associated protein 80
- UBL4A
- ubiquitin-like protein 4A.
REFERENCES
- 1. Ciccia A., Elledge S. J. (2010) The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 3. Mu J. J., Wang Y., Luo H., Leng M., Zhang J., Yang T., Besusso D., Jung S. Y., Qin J. (2007) A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J. Biol. Chem. 282, 17330–17334 [DOI] [PubMed] [Google Scholar]
- 4. Banerji J., Sands J., Strominger J. L., Spies T. (1990) A gene pair from the human major histocompatibility complex encodes large proline-rich proteins with multiple repeated motifs and a single ubiquitin-like domain. Proc. Natl. Acad. Sci. U.S.A. 87, 2374–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thress K., Henzel W., Shillinglaw W., Kornbluth S. (1998) Scythe: a novel reaper-binding apoptotic regulator. EMBO J. 17, 6135–6143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thress K., Evans E. K., Kornbluth S. (1999) Reaper-induced dissociation of a Scythe-sequestered cytochrome c-releasing activity. EMBO J. 18, 5486–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sasaki T., Gan E. C., Wakeham A., Kornbluth S., Mak T. W., Okada H. (2007) HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21, 848–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desmots F., Russell H. R., Michel D., McKinnon P. J. (2008) Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 283, 3264–3271 [DOI] [PubMed] [Google Scholar]
- 9. Wakeman T. P., Wang Q., Feng J., Wang X. F. (2012) Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J. 31, 2169–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desmots F., Russell H. R., Lee Y., Boyd K., McKinnon P. J. (2005) The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 25, 10329–10337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Minami R., Hayakawa A., Kagawa H., Yanagi Y., Yokosawa H., Kawahara H. (2010) BAG6 is essential for selective elimination of defective proteasomal substrates. J. Cell Biol. 190, 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Corduan A., Lecomte S., Martin C., Michel D., Desmots F. (2009) Sequential interplay between BAG6 and HSP70 upon heat shock. Cell Mol. Life Sci. 66, 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leznicki P., Clancy A., Schwappach B., High S. (2010) Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 123, 2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mariappan M., Li X., Stefanovic S., Sharma A., Mateja A., Keenan R. J., Hegde R. S. (2010) A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Q., Liu Y., Soetandyo N., Baek K., Hegde R., Ye Y. (2011) A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell 42, 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manchen S. T., Hubberstey A. V. (2001) Human Scythe contains a functional nuclear localization sequence and remains in the nucleus during staurosporine-induced apoptosis. Biochem. Biophys. Res. Commun. 287, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 17. Yong S. T., Wang X. F. (2012) A novel, non-apoptotic role for Scythe/BAT3: a functional switch between the pro- and anti-proliferative roles of p21 during the cell cycle. PLoS One 7, e38085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y., Qin J. (2003) MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 100, 15387–15392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Méndez J., Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zou L., Cortez D., Elledge S. J. (2002) Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chartron J. W., Suloway C. J., Zaslaver M., Clemons W. M., Jr. (2010) Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc. Natl. Acad. Sci. U.S.A. 107, 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang Y. W., Chuang Y. C., Ho Y. C., Cheng M. Y., Sun Y. J., Hsiao C. D., Wang C. (2010) Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J. Biol. Chem. 285, 9962–9970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bekker-Jensen S., Mailand N. (2011) The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 585, 2914–2919 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.