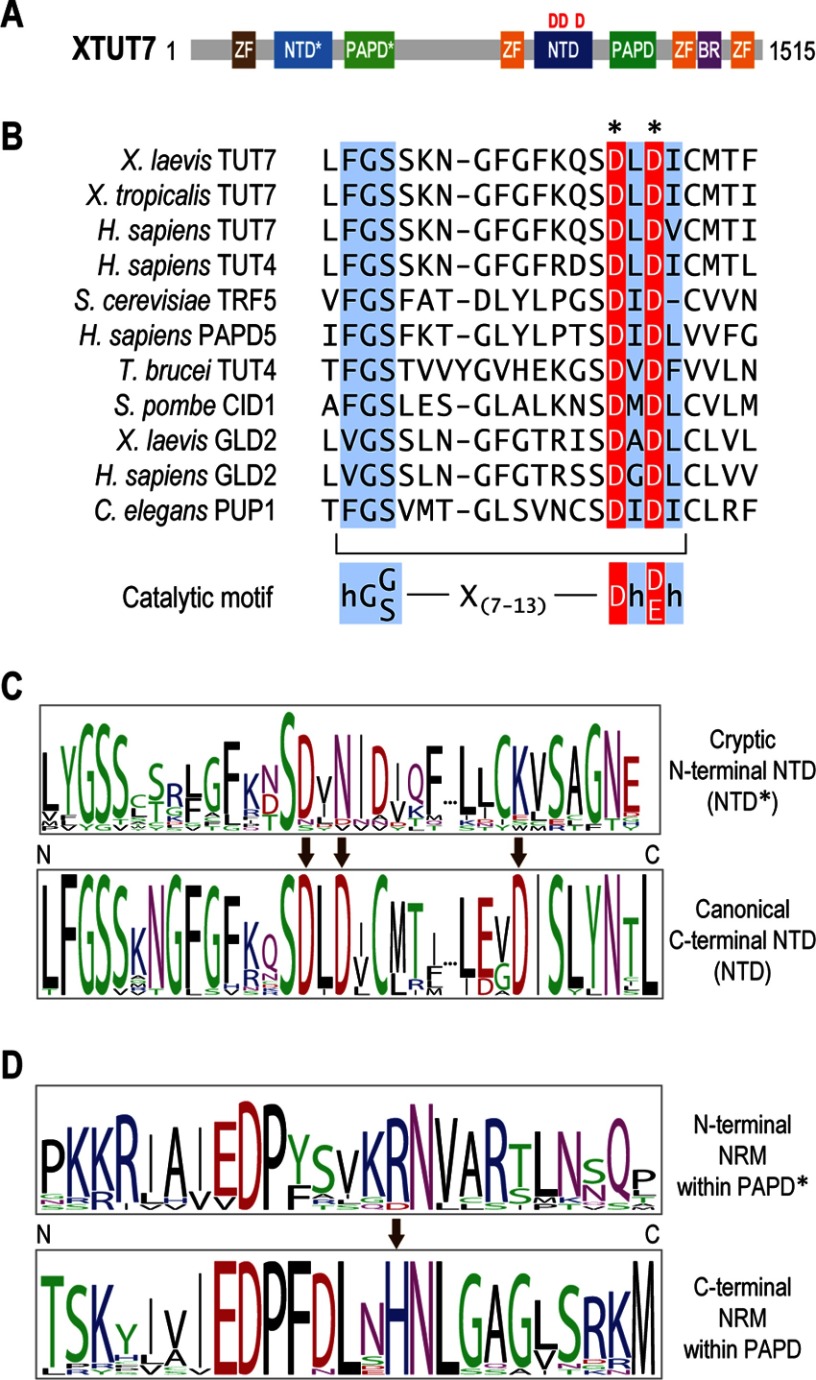
FIGURE 1.
XTUT7 domain structure. A, diagram representing XTUT7 and its predicted protein domains (amino acids indicated). Zinc finger domain (ZF) (brown), C2H2 zinc finger domain (227–252); NTD*, cryptic NTD (295–437); PAPD*, cryptic PAPD (526–575); NTD, nucleotidyl transferase domain (998–1146); PAPD, poly(A) polymerase-associated domain (1215–1268); zinc finger domain (orange), CCHC zinc finger domains (946–962, 1327–1343, 1432–1448); BR, basic region (1344–1361). The red D's denote putative catalytic aspartates. B, multiple sequence alignment of the characteristic catalytic motifs contained in the NTD of XTUT7 and other previously identified nucleotidyl transferases. The consensus rNTase catalytic motif, hG[G/S]X7–13Dh[D/E]h, is shown below the sequence alignment, where h indicates any hydrophobic amino acid (28). S. cerevisiae, Saccharomyces cerevisiae; T. brucei, Trypanosoma brucei. C, sequence logos representing multiple sequence alignments of the putative catalytic motifs contained in the NTD and NTD* of XTUT7 orthologs. The arrows indicate analogous positions in the NTD and NTD* and highlight amino acid substitutions of putative catalytic aspartates in the NTD*. D, sequence logos representing multiple sequence alignments of the NRMs encoded in the PAPDs and PAPD*s of XTUT7 orthologs. The arrow indicates an invariant histidine in the C-terminal NRM.