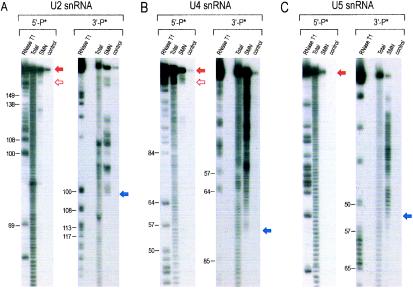
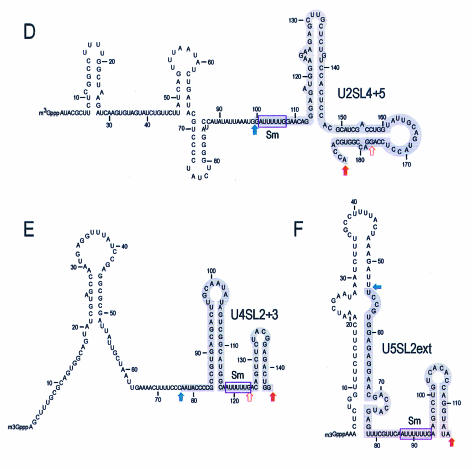
FIG. 2.
Mapping of U snRNA domains binding to the SMN complex. (A) The SMN complex-binding domain of Xenopus laevis U2 snRNA. The 5′ (5′-P*)- and 3′ (3′-P*)-end-labeled U2 snRNA was subjected to limited alkaline hydrolysis in the presence of tRNA (10 μg). The resulting hydrolyzed RNA ladders were incubated with the SMN complex. The RNA fragments bound to the SMN complex were isolated and analyzed by electrophoresis on 7 M urea-8% acrylamide gels. RNase T1-digested RNA ladders of the same RNAs were used as size markers. Solid red arrows indicate the largest extent of the SMN complex-binding domains. Open red arrows indicate the smallest possible binding fragments. Total, 5% input; control, binding in control purification. (B) The SMN complex-binding domain of chicken U4 snRNA. The same experiment was performed using 5′ and 3′-end-labeled U4 snRNAs as described for panel A. (C) The SMN complex-binding domain of X. laevis U5 snRNA. The same experiment was performed using 5′- and 3′-end-labeled U5 snRNAs as described for panel A. (D) The secondary structure of X. laevis U2 snRNA and its domain for SMN complex binding. The region denoted by the gray box (from nucleotide 100 to the 3′ end) is sufficient for the interaction with the SMN complex and is designated U2SL4 + 5. (E) The secondary structure of chicken U4 snRNAs and its domain for SMN complex binding. The region denoted by the gray box (nucleotide 77 to the 3′ end) is sufficient for the interaction with the SMN complex and is designated U4SL2 + 3. (F) The secondary structure of X. laevis U5 snRNA and its domain for SMN complex binding. The region denoted by the gray box (nucleotide 55 to the 3′ end) is sufficient for the interaction with the SMN complex and is designated U5SL2ext.