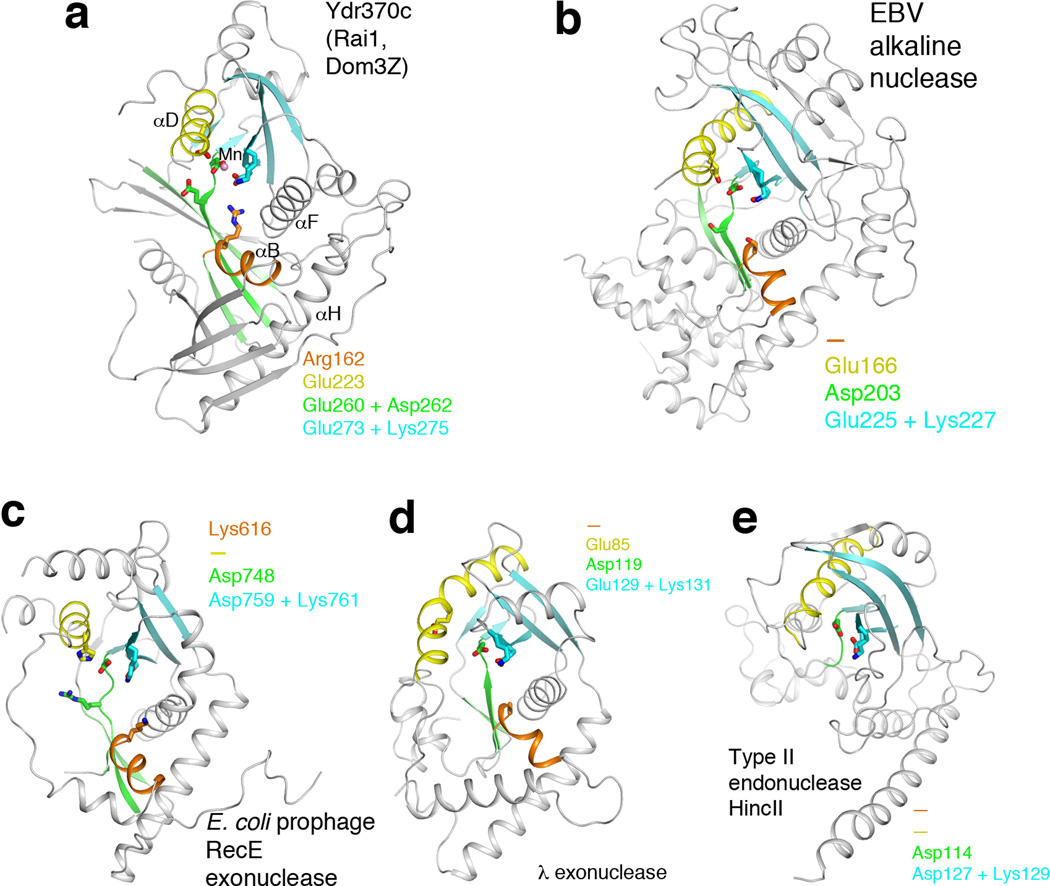
Figure 3.
Remote structural similarity to D-(D/E)XK nucleases. (a). Schematic drawing of the structure of Ydr370C. Motif I and helix αB are shown in orange, motif II and helix αD in yellow, motif III and its associated β-strands in green, motif IV and its associated β-strands in cyan. The Mn2+ ion is shown as a sphere in pink. (b). Structure of herpesvirus alkaline nuclease 19,20, in the same view as that for Ydr370C (panel a). (c). Structure of E. coli prophage RecE 5′-3′ exonuclease 22. (d). Structure of λ phage 5′-3′ exonuclease 23. (e). Structure of type II restriction endonuclease HincII 25. Residues in the four conserved motifs of Ydr370C, and their equivalents in the other enzymes, are indicated in each of the panels. A dash means that a functionally equivalent residue is absent.