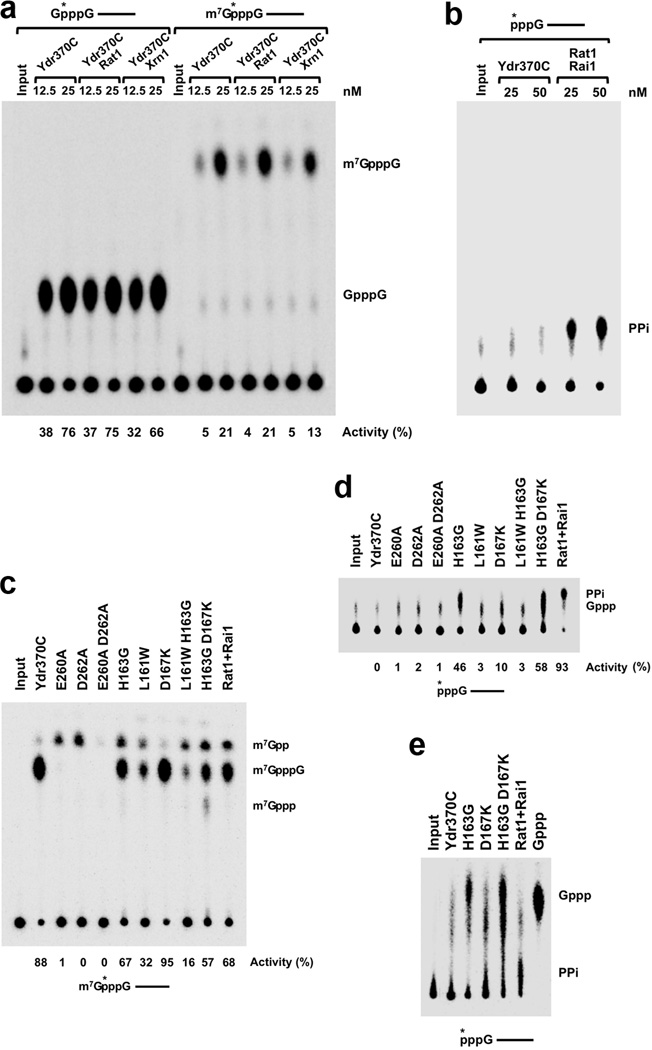
Figure 4.
Ydr370C/Dxo1 has strong decapping activity but no PPH activity. (a). Decapping assay monitored by thin-layer chromatography (TLC). Decapping activity of Ydr370C toward capped but unmethylated and mature, methylated RNA. The percentage of substrate turnover is indicated at the bottom. Rat1 and Xrn1 have no effect on the decapping activity. A schematic of the RNA used is indicated at the top with the asterisk denoting the position of the 32P-labeling. (b). PPH assay monitored by TLC. Ydr370C had essentially no PPH activity under the assay condition tested. Rai1 was used as a positive control. (c). The effects of mutations in the active site region on the decapping activity of Ydr370C. Most of the mutants were also able to release small amounts of a new product, m7Gpp. (d). PPH assay monitored by TLC. The H163G, H163G D167K, and D167K mutations in the active site region confer activity toward an RNA with 5′-end triphosphate. (e). The product released by the H163G, H163G D167K, and D167K mutants from an RNA with 5′-end triphosphate is GTP (Gppp), rather than pyrophosphate (PPi). A different running buffer was used to more clearly separate PPi from Gppp. Gppp in the sample lanes migrates slightly differently compared to the marker, as the samples are in a buffer and also contain protein while the marker is in water.