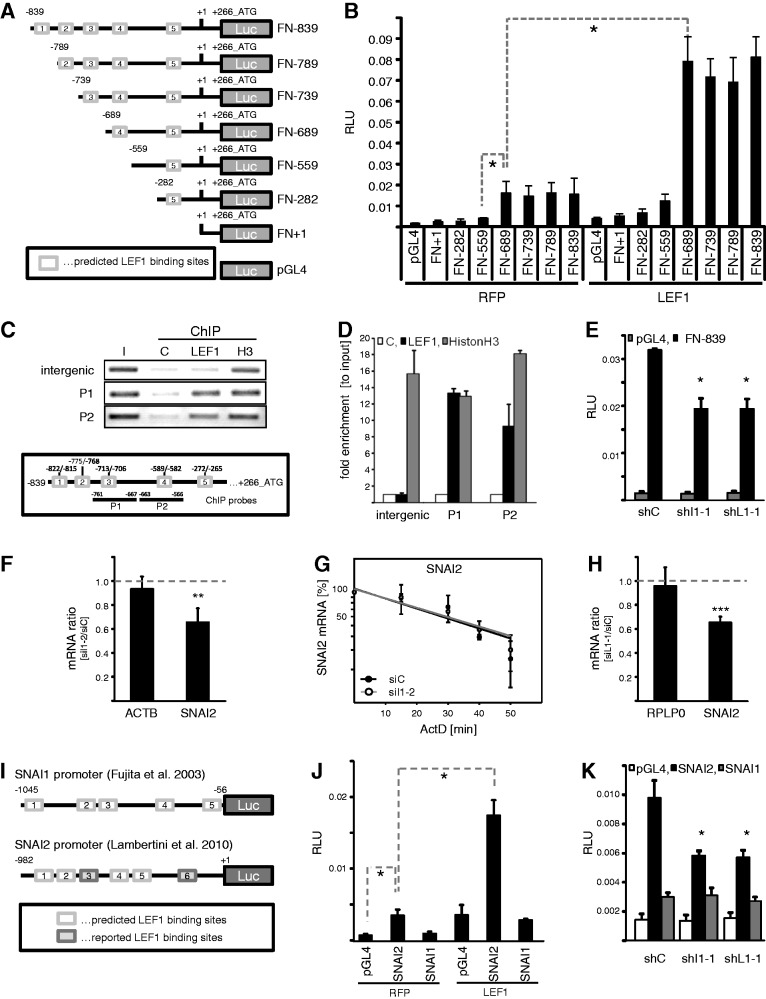
Figure 5.
IGF2BP1 modulates FN1 and SNAI2 (SLUG) transcription via LEF1. (A) Schematic of luciferase reporters comprising the full-length in silico predicted (FN-839) or 5′-truncated fragments of the human FN1 promoter. The proposed transcription start is indicated by +1 with a reported 5′-UTR of 266 nt. Putative LEF1-binding sites predicted by ‘PROMO’ are depicted as white boxes with labels ‘1-5’ in 5′-to-3′ direction. (B) The Firefly luciferase activity of indicated promoter fragments or empty pGL4 vector was monitored in HEK293 cells on transient co-transfection with RFP or LEF1 for 30 h. Firefly activities were normalized by Renilla activities [relative luciferase units (RLU)], serving as internal controls. All reporters comprising the putative LEF1-binding site four showed promoter activity and were activated by LEF1. (C and D) Binding of endogenous LEF1 protein to the human FN1 promoter in HEK293 cells was assessed by ChIP. The association of endogenous LEF1 or histone H3 to the FN1 promoter was monitored by semi-quantitative (C) as well as quantitative PCR (D) using to FN1 promoter specific amplicons (P1 and P2, indicated in lower panel). An intergenic probe served as positive control. IgG-agarose was used to monitor unspecific binding (C, negative control). In (D), the enrichment of indicated genomic DNA fragments (P1 and P2) or the intergenic control (intergenic) was determined relative to the diluted input fraction (I) normalized by IgG-controls using the ΔCt-method. (E) HEK293 cells were co-transfected with FN-839 luciferase reporter and IGF2BP1-directed (shI1-1), LEF1-directed (shL1-1) or control shRNA encoding vectors for 48 h. RLUs were determined as described in (B). (F) HEK293 cells were transfected with IGF2BP1-directed (siI1-2) or control siRNAs (siC) for 72 h. The abundance of SNAI2 mRNA in response to IGF2BP1 knockdown was analyzed by qRT-PCR using the ΔΔCt-method and PPIA for normalization. ACTB served as control. (G) HEK293 cells transfected as in (F) were treated with ActD (5 µM) to block transcription for indicated times. SNAI2 mRNA turnover was analyzed by qRT-PCR using the ΔΔCt-method and PPIA for normalization. RNA decay is depicted in semi-logarithmic scale revealing no significant difference in mRNA turnover (P-value not shown). (H) HEK293 cells were transfected with LEF1-directed (siL1-1) or control siRNAs (siC) for 72 h. The abundance of SNAI2 mRNA in response to LEF1 depletion was analyzed by qRT-PCR using the ΔΔCt-method and PPIA for normalization. RPLP0 served as control. (I) Schematic of Firefly luciferase reporters comprising the SNAI1 or SNAI2 promoter sequences, as previously reported (37,45). Indicated putative LEF1-binding sites within the SNAI1 or SNAI2 promoter were predicted [white boxes; as described in (A)] or as previously reported [gray boxes, only for SNAI2; (37)]. (J) The Firefly activity of SNAI1 or SNAI2 promoter fragments cloned in pGL4 as well as the activity of empty pGL4 vector was monitored in HEK293 cells on transient co-transfection with RFP or LEF1 for 30 h. RLUs were determined as described in (B). LEF1 only enhanced the activity of the SNAI2 promoter. (K) HEK293 cells were co-transfected with SNAI1 or SNAI2 promoter reporters and indicated shRNA-encoding vectors for 48 h. RLUs were determined as described in (B). SNAI2 promoter activity was reduced by IGF2BP1 as well as LEF1 knockdown, whereas the SNAI1 reporter activity remained largely unaffected and was barely elevated compared with the empty control reporter. Statistical significance was validated by Student’s t-testing: *P < 0.05; ***P < 0.0005. Error bars indicate SD of at least three independent analyses.