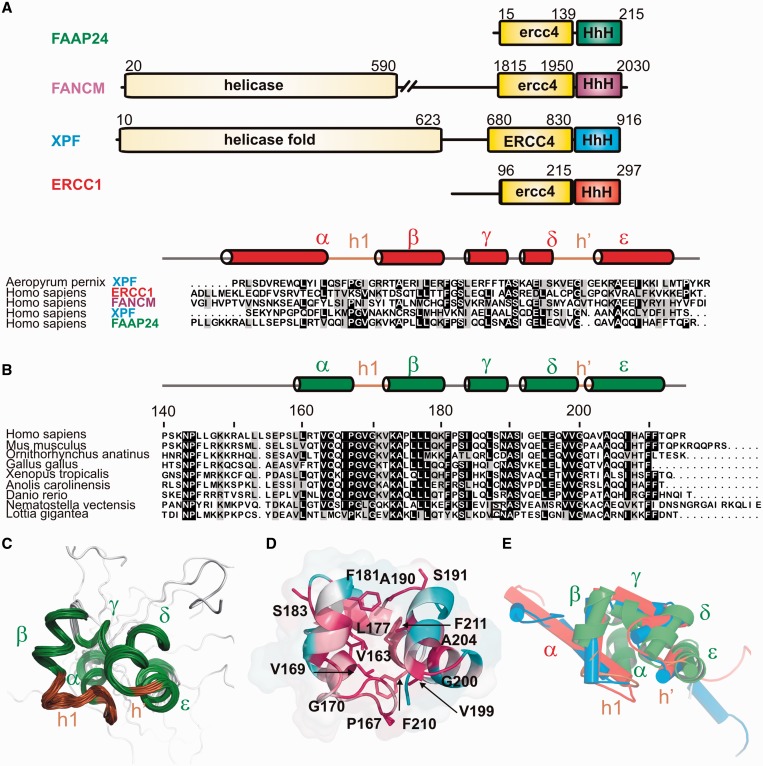
Figure 1.
Solution structure of the HhH domain of FAAP24. (A) Domain organization of FAAP24 (with the HhH domain structure in green), FANCM (purple), XPF (blue) and ERCC1 (red). ERCC4 refers to the catalytically active nuclease domain; ercc4 refers to the structurally homologous domain that lacks endonuclease activity or nuclease signature. The lower panel shows the structure-based alignment of the HhH domain sequences colored using the Boxshade server; for comparison, an archaeal XPF sequence is depicted. The secondary structure elements are indicated for the ERCC1 HhH domain [1z00 (16)], the hairpin regions (h1, h′) are depicted in brown. (B) A representative multiple sequence alignment of the FAAP24 protein family (140–215) made using clustalX (17), showing the most distantly related FAAP24 HhH orthologous sequences obtained from the OMA database (18). Coloring is based on all orthologues and performed with Boxshade with a shading threshold at 0.8. Secondary structure elements for FAAP24 HhH domain are depicted above, h’ refers to the second HhH motif that lacks a classical hairpin region. (C) Ensemble of the 20 lowest energy structures of the FAAP24 HhH domain (140–215). Secondary structure elements are depicted in green, the hairpin domain regions brown. (D) Cartoon representation of the FAAP24 HhH domain structure colored according to sequence conservation, least conserved residues are colored turquoise and most conserved red. The latter residues are depicted in a stick representation. Sequence conservation was calculated using Consurf (19) with the multiple sequence alignment from 1B. The N-terminal tail (140–157) of FAAP24 is structurally disordered and omitted for clarity in most representations. (E) Overlay of the HhH domain structures of FAAP24 (green), ERCC1 (red; 1z00), XPF (blue; 1z00) with helices presented as cylinders, the FAAP24 HhH domain structure is also shown in green in cartoon representation.