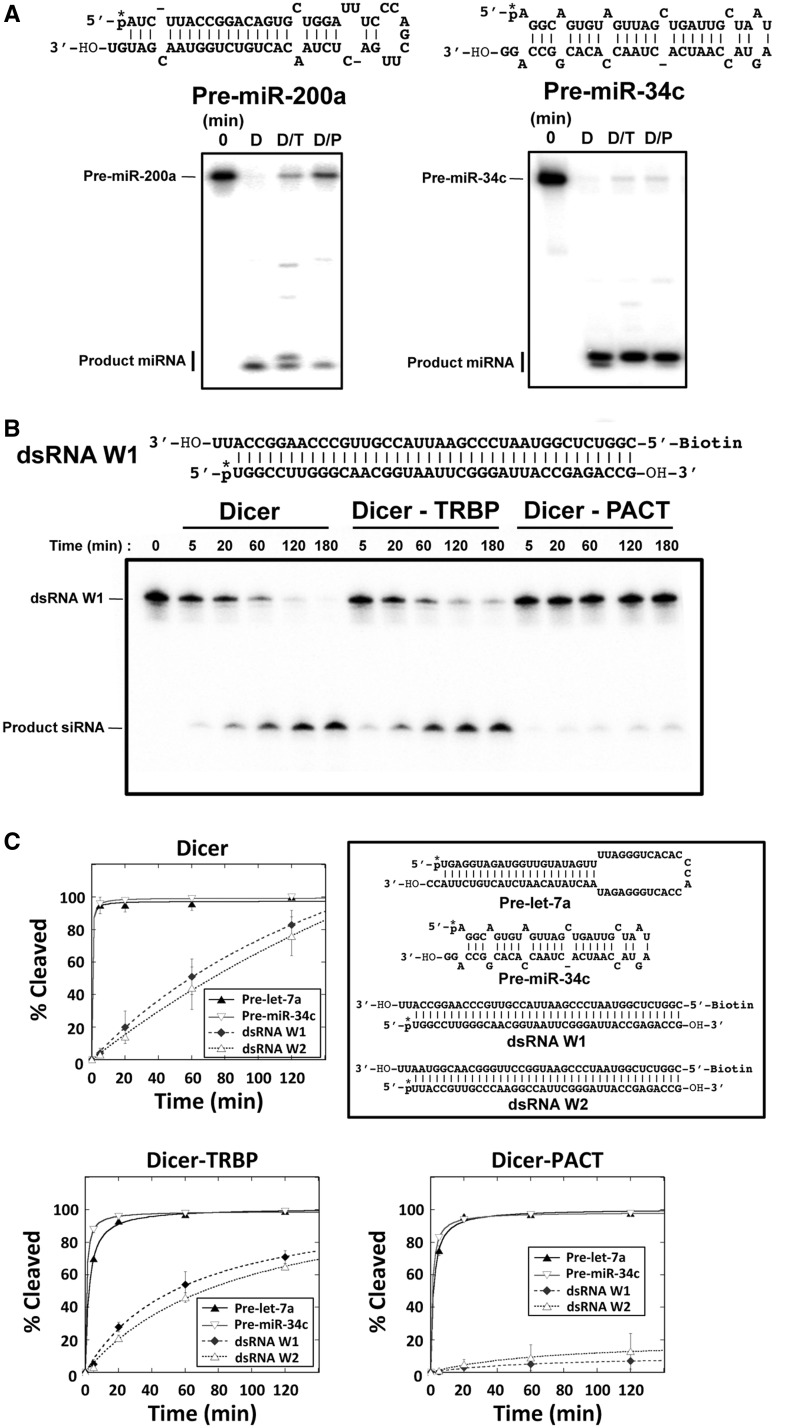
Figure 2.
TRBP and PACT affect pre-miRNA and pre-siRNA processing by Dicer in non-redundant fashion. (A) Dicer–TRBP (D/T) and Dicer–PACT (D/P) are not redundant in producing different-sized miRNAs in pre-miR-200a and pre-miR-34c processing. Assays were done in a single turnover condition with 10-fold excess amount of protein/complex to RNA. RNA substrates were labeled by 32P phosphate at 5′ end, which is marked with an asterisk. [RNA] = 5 nM and [Dicer]/[Dicer–TRBP]/[Dicer–PACT] = 50 nM were incubated in reaction buffer for 60 min (see ‘Materials and Methods’ section) and quenched with 1.2 volumes of 2-fold formamide dye. Samples were boiled at 70°C for 10 min before loading to 12% denaturing PAGE. After drying, the gel was exposed to a phosphoscreen. (B) Dicer–PACT processes pre-siRNA (dsRNA W1) much less efficiently than Dicer and Dicer–TRBP in a single turnover condition with 10-fold excess amount of protein/complex to RNA. dsRNA W1 was labeled with 32P phosphate at 5′ end of the bottom strand, which is marked with asterisk. [RNA] = 5 nM and [Dicer]/[Dicer–TRBP]/[Dicer–PACT] = 50 nM were incubated in reaction buffer. The reactions were quenched at each time point and the following steps were the same as described in (A). (C) Dicer, Dicer–TRBP and Dicer–PACT processing kinetics for pre-miRNAs (pre-let-7a and pre-miR-34c) and pre-siRNAs (dsRNA W1 and dsRNA W2). Dicer–PACT processes pre-siRNA much slower than Dicer and Dicer–TRBP, while processing pre-let-7a and pre-miR-34c as efficiently as Dicer and Dicer–TRBP. A single turnover reaction condition with 10-fold excess amount of protein/complex to RNA was used for kinetic assays as in (A). The quenched samples were boiled and loaded onto 12% denaturing gel. After drying and exposing the gel to phosphoscreen, precursor substrate and product miRNA or siRNA bands were quantified by ImageQuant software (GE Healthcare life sciences). % Cleavage was calculated by 100 × (product miRNA counts)/(total counts − sum of substrate and product RNA counts).