Figure 4.
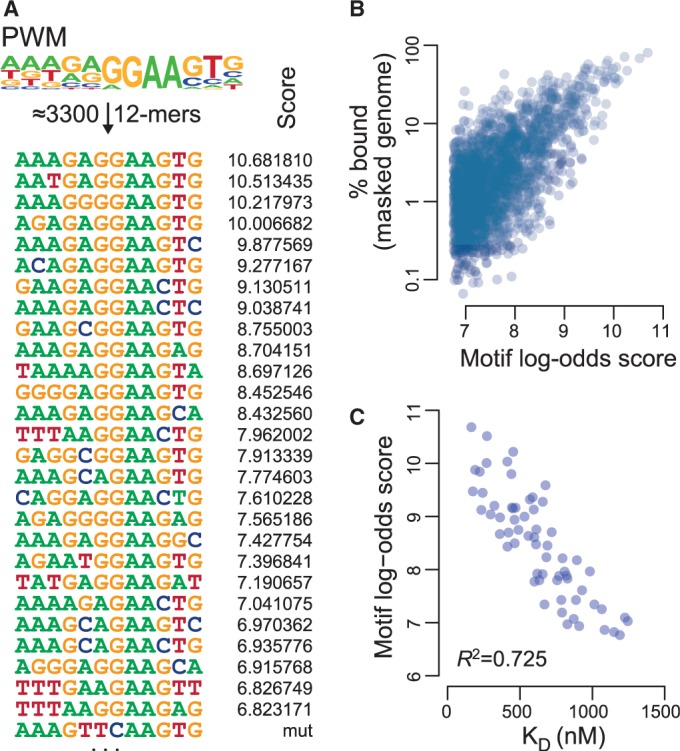
Motif scores correlate with in vitro binding affinity. (A) The ChIP-seq–derived PU.1 consensus PWM (sequence logo is shown on top) comprises ∼3300 different 12mers. Twenty-five representative 12mers overlapping the PWM are shown below the logo next to their motif log-odds scores. (B) Scatter plot showing scores of motifs relative to the frequency of in vivo binding events across the repeat-masked genome (only motifs with >100 occurrences across the masked genome are included). (C) Scatter plot showing the correlation between microscale thermophoresis-derived dissociation constants (KD-values) for the interaction between recombinant full-length PU.1 and double-stranded oligonucleotides [including the ones listed in (A)] with corresponding motif log-odds scores. Only KD-values <1500 mM were plotted. Oligos with larger KD-values (between 1500 and 4500 mM, 12 in total) had low-log-odds scores (between 6.7 and 7.6). A complete list of KD- and log-odds score values is given in Supplementary Table S1.
