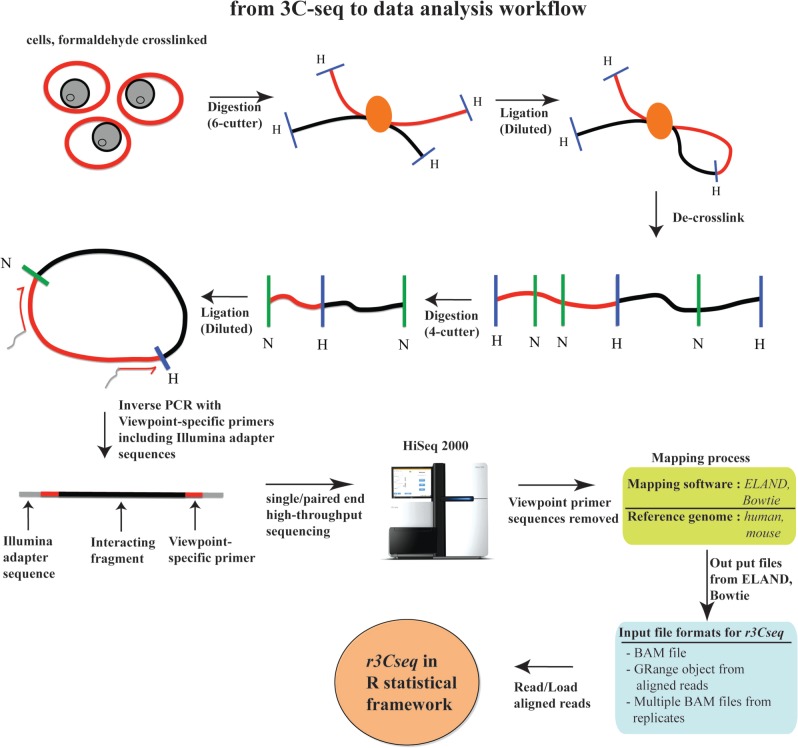
Figure 1.
3C-seq experimental procedures and data analysis workflow. Formaldehyde cross-linked chromatin is digested with a six-cutter restriction enzyme and ligated under dilute conditions. After de-cross-linking, DNA is digested with a four-cutter enzyme and again ligated under dilute conditions to create small circular fragments representing individual ligation events. Inverse PCR using viewpoint-specific primers containing Illumina sequencing adapters is used to generate a viewpoint-specific 3C-seq library. After high-throughput sequencing, reads are trimmed and mapped to the reference genome, after which they are loaded into the r3Cseq software.