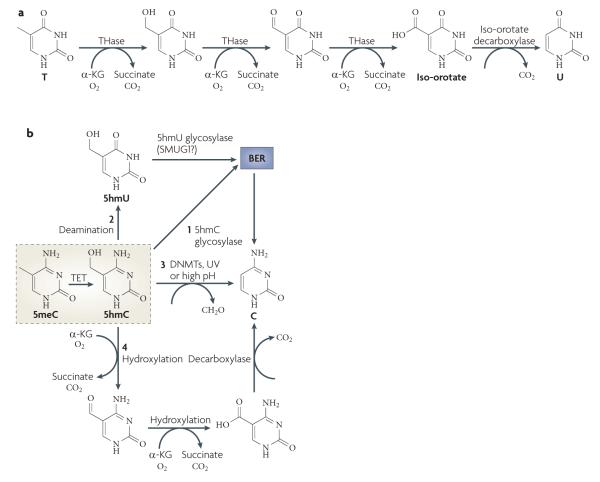
Figure 5. Oxidative demethylation by TET proteins.
a | Part of the thymidine salvage pathway. Direct removal of the methyl group of 5-methylcytosine (5meC) involves breaking a carbon-carbon bond, which requires an enzyme with great catalytic power. Such an enzyme exists in the thymidine salvage pathway. Starting with T, thymine-7-hydroxylase (THase) carries out three consecutive hydroxylation reactions to produce iso-orotate, which is processed by a decarboxylase to produce U. A similar mechanism may be used in active DNA demethylation, particularly by the ten-eleven translocation (TET) family of proteins. b | The fate of 5-hydroxymethylcytosine (5hmC). The TET family of proteins catalyses the conversion of 5meC to 5hmC, which may be an intermediate that can be further processed by one of the following mechanisms. BER may be initiated by a 5hmC glycosylase (1); 5hmC may undergo deamination to produce 5hmU (2), which is repaired by BER through a 5hmU glycosylase such as SMUG1 (single-strand-selective monofunctional U DNA glycosylase 1); 5hmC may directly be converted to C by DNA methyltransferases (DNMTs), ultraviolet (UV) exposure or high pH (3); or consecutive hydroxylation reactions followed by a decarboxylation reaction similar to the thymidine salvage pathway may be used to ultimately replace 5hmC with C (4). Alternatively, 5hmC itself may be a functional modification. α-KG, α-ketoglutarate.